Figure 4.
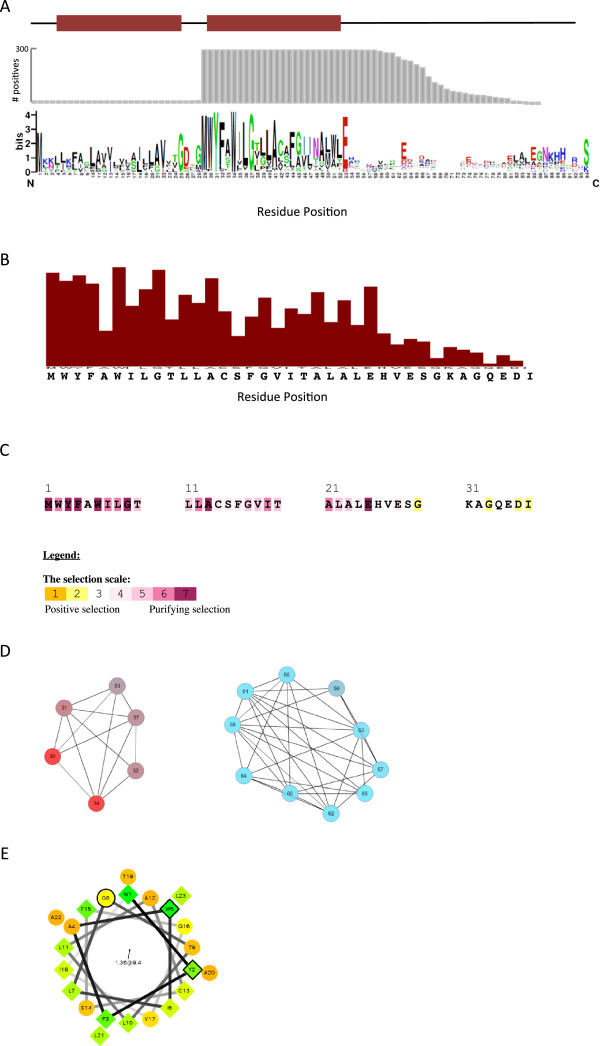
Sequence analysis of the CydX protein family. (A) Consensus sequence of CydX homologues compared to the presence of predicted transmembrane domains (red bars) and the number of homologues that contain amino acids at each position (grey bars). The sequence logo was created using a MUSCLE alignment [57] analyzed by the WebLogo program [57]. Amino acids are colored based on their properties at physiological conditions as follows: black amino acids are hydrophobic, green residues are hydrophilic, blue residues are positively-charged and red residues are negatively-charged. Transmembrane domains were predicted using the program TMHMM [56]. (B) Predicted evolutionary importance of each residue in CydX. Analysis performed using the Lichtarge Computational Biology Lab’s Universal Evolutionary Trace web server [57]. (C) Predicted selection pressure on each amino acid in the CydX protein. Analysis performed using the Selecton program. (D) Residues within the CydX protein that share mutual information. Analysis performed using the MISTIC program. Residues are colored based on conservation, with the amino acids in red positions in the alignment being conserved and blue amino acids showing less conservation. (E) Alpha-helical wheel project of the predicted transmembrane domain of the E. coli CydX protein [28]. The conserved residues Y3, W6 and G9 are outlined in black. The shapes the amino acids are based on their properties at physiological conditions as follows: hydrophobic residues are diamonds and hydrophilic residues are circles. The degree of hydrophobicity of diamond residues is also reflected in the color, with green being most hydrophobic and yellow being least hydrophobic, and a range of color between those depending on predicted hydrophobicity. Likewise, the degree of hydrophilicity of circle residues is reflected in the color, with red being most hydrophilic and light orange being least, and a range of color between those depending on predicted hydrophilicity.
