Figure 9.
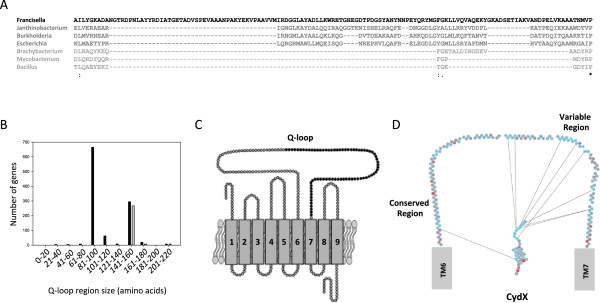
Synteny between the cydX gene and the long Q-loop allele of cydA . (A) Alignment of the Q-loop region from select CydA homologues. Sequences are shaded in a gradient going from longest Q-loop (darkest) to shortest Q-loop (lightest). (B) Histogram showing the number of CydA homologues containing Q-loops of increasing size (black bars) and the number of CydA proteins encoded in an operon also containing cydX (grey bars). (C) Diagram of the CydA protein containing the Q-loop. Residues shown in black are those that are present only in long Q-loop CydA variants. (D) Diagram showing mutual information shared between residues in the CydX protein, shown in its predicted orientation in the inner membrane, and the Q-loop, shown as the residues spanning transmembrane regions 6 (TM6) and 7 (TM7) of CydA. Lines between residues show high mutual information between residues. The conserved and variable regions of the Q-loop are labeled. Spaces between residues in the Q-loop region represent residues that are missing because they either show no mutual information or share mutual information with other Q-loop residues and not with CydX. A mutual information filter cutoff of 10 was used for this figure. Species are as follows: Francisella philomiragia subsp. philomiragia ATCC 25017 (“Francisella”), Janthinobacterium sp. Marseille (“Janthinobacterium”), Burkholderia xenovorans LB400 (“Burkholderia”), Escherichia coli 536 (“Escherichia”), Brachybacterium faecium DSM 4810 (“Brachybacterium”), Mycobacterium marinum M (“Mycobacterium”), and Bacillus subtilis subsp. spizizenii str. W23 (“Bacillus”). Mutual information was determined using the program MISTIC. Alignments were generated using the program MUSCLE [57]. ‘*’ indicates that the residues are identical in all sequences and ‘:’ and ‘.’, respectively, indicated conserved and semi-conserved substitutions as defined by MUSCLE.
