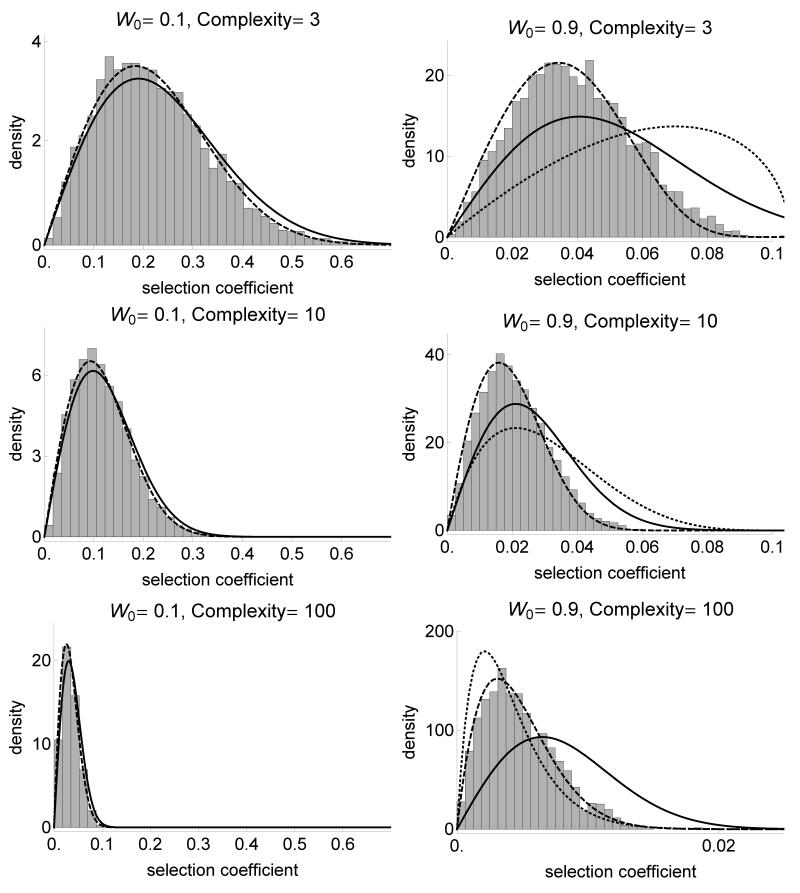
Figure 2.
Distribution of the selection coefficient under Fisher’s model for various fitnesses of the ancestral strain W0 (left: far from the optimum, right: near the optimum) and various complexities of the phenotypic space (from bottom to top). The line is the χ2 analytical approximation (equation 2), the dashed line is based on the gamma approximation developed in Martin and Lenormand (2006) (Appendix) and the dotted line for W0 = 0.9 is the beta approximation developed in Martin and Lenormand (2008). Selection coefficient is calculated for 10000 selected mutations. σmut is scaled such that the average norm of the mutational effect on phenotype is constant equal to 0.1 across complexities. The population size is N = 107 and the mutation rate μ = 10−9.