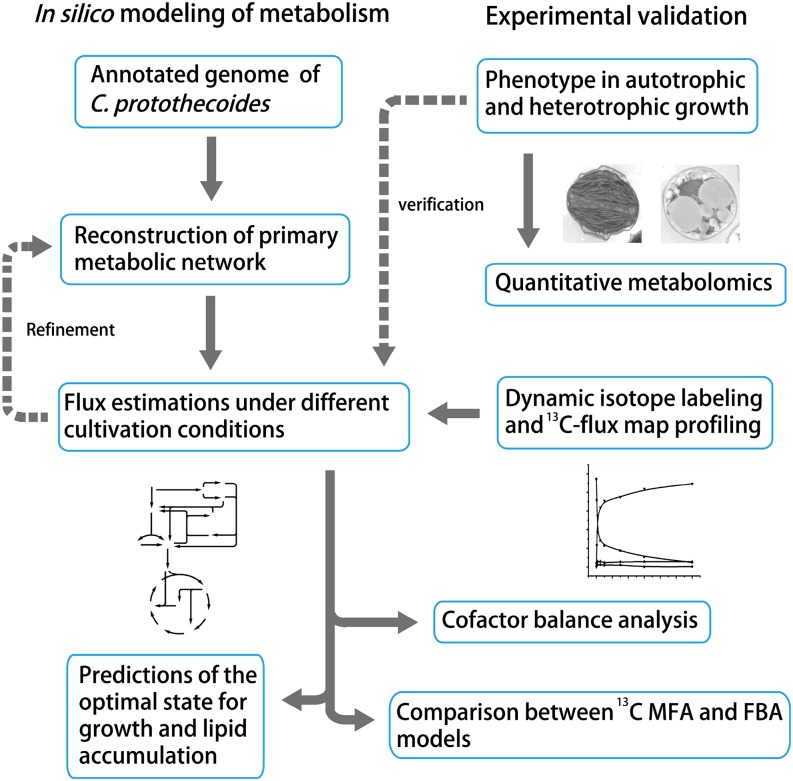
Figure 1.
Schematic MFA workflow of C. protothecoides using computational and experimental approaches. Briefly, we reconstructed the primary metabolic network of the oleaginous microalga C. protothecoides based on the recently available gene functional annotation (Gao et al., 2014). Intracellular fluxes under different cultivation conditions were simulated, and the optimal solutions were predicted using cell growth and lipid accumulation as the objective functions. The genome-based metabolic model was further constrained by experimental metabolomics and dynamic isotope labeling for INST-MFA (Young, 2014). Results from 13C MFA measurement and FBA prediction are comparably analyzed.