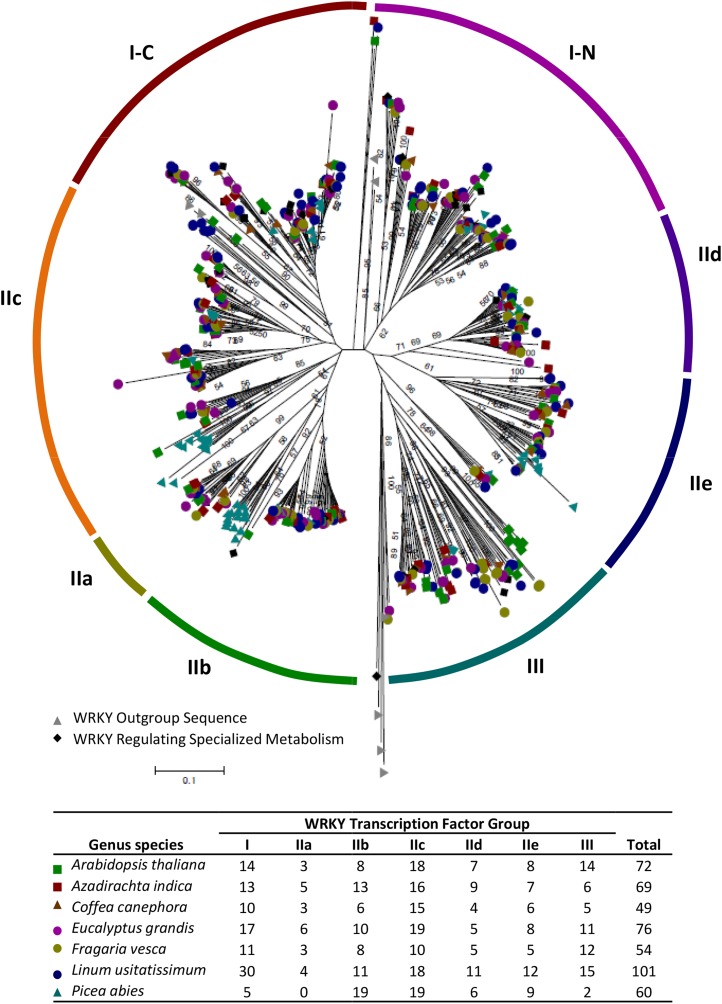
Figure 2.
Phylogenetic analysis of Arabidopsis (green squares), coffee (brown triangles), eucalyptus (purple circles), flax (blue circles), neem (red squares), spruce (teal triangles), and strawberry (gold circles) WRKY families alongside those regulating specialized metabolism in other species (top) and the number of TFs present in each group (bottom). Human GCMa and FLYWCH plus G. lamblia, D. discoideum, and C. reinhardtii WRKYs were used to form an outgroup to root the phylogenetic tree (gray triangles). WRKY TFs regulating specialized metabolites are denoted with black diamonds. The neighbor-joining tree was constructed using the proportion of amino acids differing between sequences (i.e. the p-distance method) and 1,000 bootstrap replications using MEGA 6.0. Sequence alignment was performed using the ClustalW algorithm. The phylogenetic tree has been deposited into TreeBase under identification number 16440.