FIGURE 1.
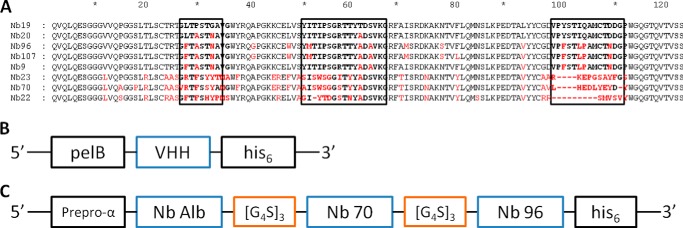
A–C, primary structure of eight different anti-hTNFR1 nanobodies, the gene construct in the pHEN6c vector of monovalent nanobodies, and the pAOXZalfa vector of TROS. A, amino acid sequences of the eight nanobodies. The CDRs are shown in boldface in boxes. Amino acids in red indicate the main differences between the different Nbs and Nb 19 (top sequence). They belonged to four different clonally unrelated B-cell clones. The amino acid sequences suggest that Nbs 19, 20, 96, 107, and 9 are from clonally related B-cells resulting from somatic hypermutations. Nb 23, 70, and 22 belong to unrelated B-cell clones. Gaps (dashes) are introduced to align sequences. B, this pHEN6c construct for the Nanobody was transformed in WK6 E. coli cells. The VHH gene of the Nb is preceded by the pelB leader signal sequence, which directs the expressed protein to the bacterial periplasm and ends with a sequence encoding a C-terminal His6 tag. C, in the gene construct of TROS, Nb Alb, Nb 70, and Nb 96 are linked to each other with the flexible (G4-S)3 linker. The TROS construct is preceded by the α-mating factor pre-pro signal sequence and is made for expression in the yeast P. pastoris.
