FIGURE 4.
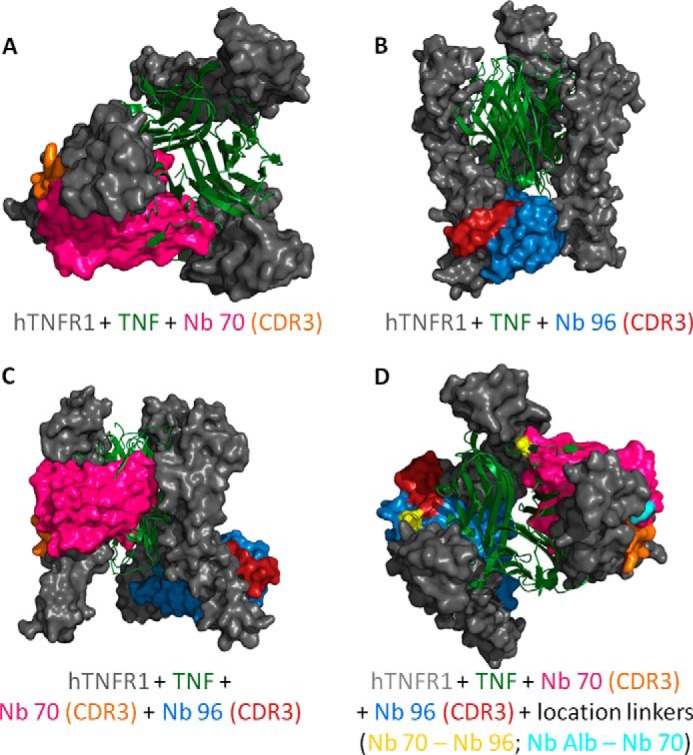
A–D, docking prediction models of trimeric hTNFR1 interacting with TNF, Nb 70, and Nb 96. A and B, Nb 70 competes with TNF (top view), whereas Nb 96 does not (frontal view). Their CDR3 binds into smaller clefts of the receptor and is important for the high affinity. C, prediction model of Nb 70 and Nb 96 interacting simultaneously with hTNFR1. They interact with the three-dimensional structure of hTNFR1, as their binding with the different chains of trimeric hTNFR1 is not interchangeable. D, generation of a trivalent Nanobody by linking the C-terminal end of an anti-albumin Nb to the N-terminal end of Nb 70 and its C-terminal end to the N-terminal end of Nb 96 might increase the inhibitory capacity of the Nb. The anti-albumin Nb does not disturb the binding of Nb 70 or Nb 96 to hTNFR1, as this linker is located on the outside (start linker light blue), although the flexible linker between Nb 70 and Nb 96 (start/end linker yellow) goes through the different chains of trimeric hTNFR1, thereby disturbing its interaction with TNF even more. All models were validated by RAMPAGE (29), and the best models were used for docking by ClusPro (30) to predict binding of Nb 70 and Nb 96 to hTNFR1. Homology models and docking results were analyzed and figures rendered using PyMOL.
