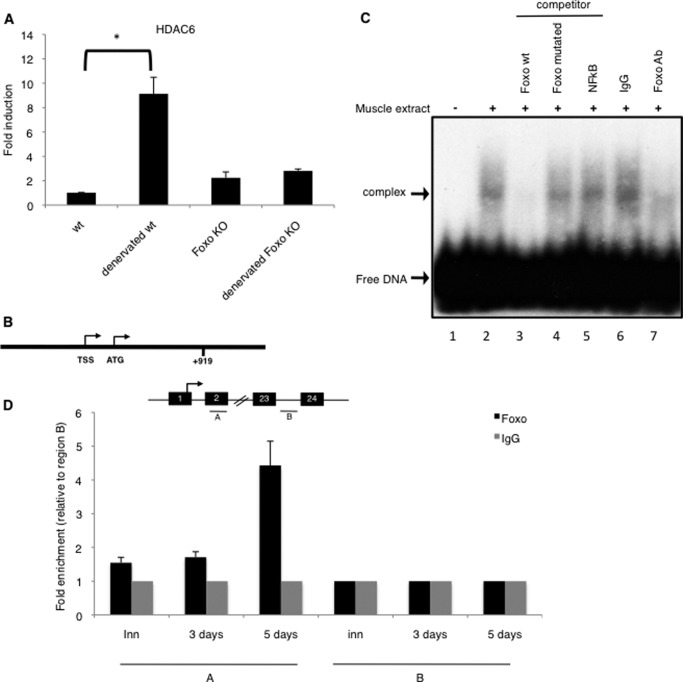
FIGURE 3.
A, up-regulation of HDAC6 in FoxO triple KO and wild type mice TA muscles after 5 days of denervation. QPCR analysis was performed on 3 mice for each condition. Results are presented as fold induction compared with innervated. Error bars represent S.E. The asterisk indicates statistically significant changes at p < 0.01 (p = 0.004). B, consensus sequence for FoxO3 (GTAAAGA) at position +919 in the HDAC6 gene region. C, muscle extracts from denervated TA muscles were tested for binding to double stranded 32P-labeled oligonucleotides containing a FoxO3 binding site by EMSA. Arrows indicate the FoxO3-DNA complex and the free DNA. Lane 1: free DNA alone; all other lanes contain the protein extract and, as indicated at the top of the figure, no oligonucleotide (positive control; lane 2), a competitor oligonucleotide (FoxO wt, FoxO mutant or NFkB; lanes 3–5) or an antibody (IgG or anti-FoxO3; lanes 6–7). D and E, localization of FoxO3 protein at the HDAC6 gene. Chromatin immunoprecipitation (ChIP) analysis were performed in wild type mice TAs muscles, either innervated, or 3 or 5 days denervated. Two indicated regions (A and B) were tested for QPCR amplification. QPCR values were calculated by extrapolation from a standard curve of Inputs DNA dilutions. Enrichment values were normalized to IgG signals and shown as the fold difference relative to region B. Each ChIP-QPCR histogram indicates the mean of triplicate results, error bars indicate S.E. A: FoxO binding site region at +919, B: negative region at +15074.