FIGURE 3.
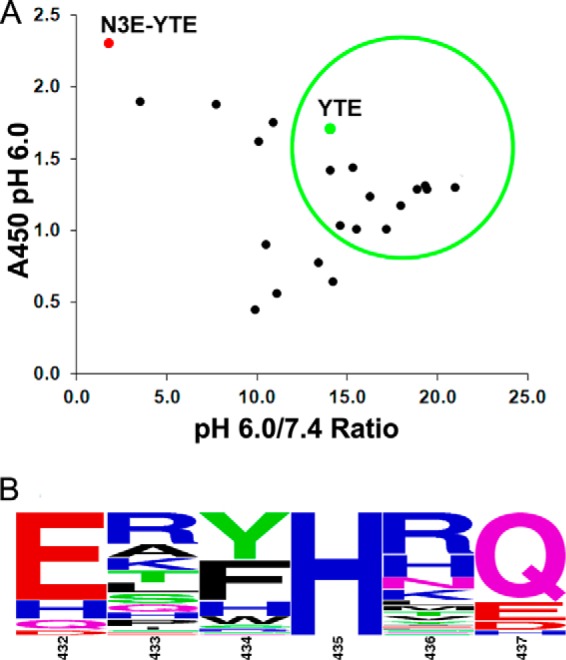
Sequence analysis and phage ELISA results of variants identified from the charged library. A, phage ELISA data are presented as shown in Fig. 2. The majority of variants identified from the charged library show high affinity and pH-dependent binding. Clones displayed in the green circle were converted to full-length IgG and reassessed for FcRn binding (see Table 1). B, sequence logo representation of a total of 68 phage clones identified after four rounds of phage panning. The relative letter size represents the frequency of the amino acid occurring at the position. Positions 433, 434, 436, and 437 are randomized in the library. Amino acids Glu, Asp, His, or Gln are encoded in positions 432 and 437. His-435 was not mutated in the library.
