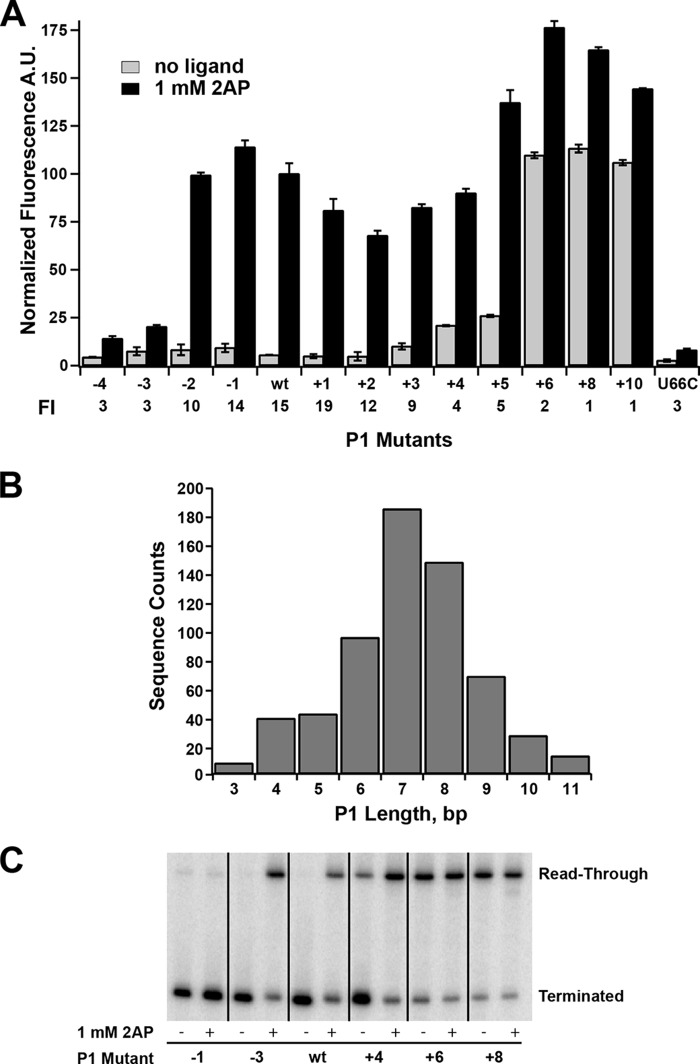
FIGURE 4.
Effect of mutations that change the length of the P1 helix on regulatory activity. A, effect of point mutations on the 5′-side of P1 that either remove (−4 through −1) or add (+1 through +10) Watson-Crick pairs on 2-AP-dependent regulatory activity. A binding-incompetent mutant (U66C) is shown for comparison. The -fold induction (FI) values are given below the graph. Error bars represent the S.E. of three separate experiments. A.U., arbitrary units. B, histogram of the length distributions of the P1 helix found within the purine riboswitch family using all non-redundant sequences deposited in Rfam 11.0 (33). This distribution corresponds to an analysis considering the P1 helix to be strictly a contiguous stretch of canonical base pairs (A-U, G-C, and G·U). C, single-turnover transcription assays of a subset of P1 length variants tested in A. The values for the percentage read-through transcription in the absence and presence of 1 mm 2-AP are given in Table 3.