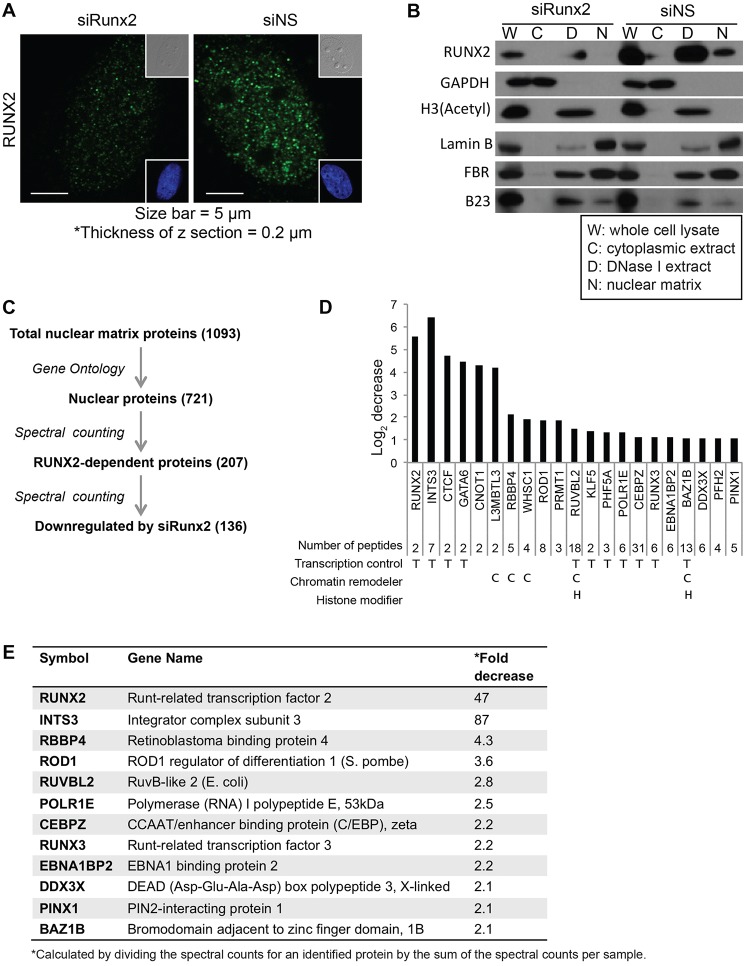
Fig. 1.
Proteomic analysis of RUNX2-related nuclear matrix proteins in RUNX2-knockdown cells. (A) Immunofluorescence staining of Saos2 cells transfected with either nontargeting siRNA (siNS) or RUNX2-targeting siRNA (siRUNX2). Insets show differential interference contrast (DIC, upper right) and DAPI images (lower right). (B) Proteins in whole-cell lysates, cytoplasmic extracts, DNase I/salt extracts and nuclear matrix fractions, were resolved by 15% SDS-PAGE and analyzed by western blotting using primary antibodies specific for the indicated proteins. GAPDH, histone H3 and lamin B were used as markers for cytoplasmic extracts, DNase I/salt extracts and nuclear matrix fractions, respectively. FBR (fibrillarin) and B23 (nucleophosmin) are nuclear matrix components that are expected to be recovered in both DNase I/salt extracts and the nuclear matrix fraction. (C) Workflow for the proteomic screening of RUNX2-dependent nuclear matrix proteins. A total of 1093 proteins were identified by mass-spectrometry-assisted fingerprinting of the nuclear matrix fraction prepared from Saos2 cells transfected with nontargeting siRNA or RUNX2-targeting siRNA. Of these, 721 were identified as nuclear proteins by Gene Ontology (GO) analysis. Spectral counting obtained from mass spectrometry was used to compare the relative fold-change of protein levels; 136 proteins out of 207 RUNX2-dependent nuclear proteins were identified as nuclear proteins that were downregulated by RUNX2 knockdown (see supplementary material Table S1). A functional subset of proteins, including chromatin remodelers, epigenetic regulators, transcriptional controllers and most RUNX2-dependent nuclear proteins, was selected by further screening for proteins identified through ≥5 peptides (supplementary material Table S2). (D) The graph shows the log (base 2) fold-change in protein levels between nuclear matrix fractions of cells transfected with RUNX2-targeting siRNA or nontargeting siRNA determined by spectral counting obtained from mass spectrometry analysis. Results for a functional subset of proteins, including transcription regulators, chromatin remodelers and histone modifiers is shown. The number of unique peptides identified by mass spectrometry is indicated, as are the biological functions of proteins – T, transcription regulator; C, chromatin remodeler; H, histone modifier. (E) Functions and fold decreases due to RUNX2 knockdown of representative proteins from the mass spectrometry analysis of nuclear matrix proteins from siRNA-transfected Saos2 cells. The fold decrease in protein levels was calculated by dividing the spectral counts for an identified protein by the sum of the spectral counts per sample.