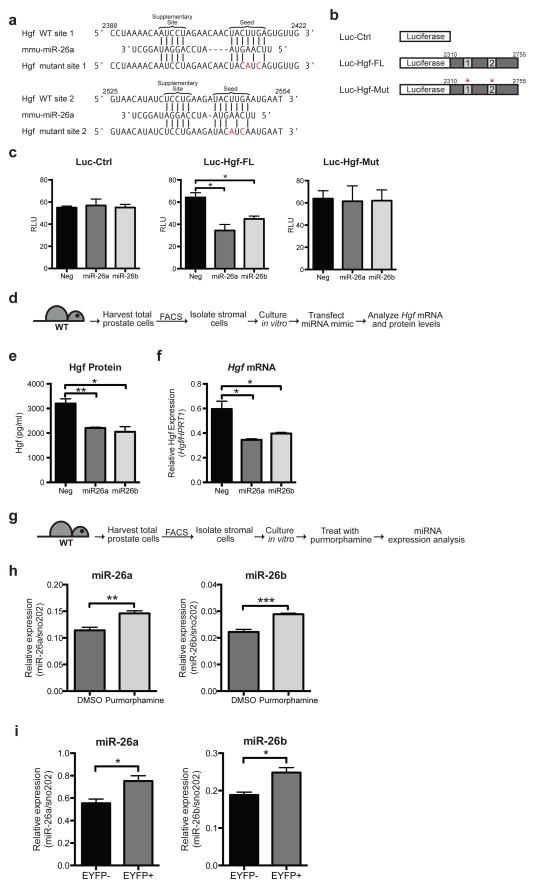
Figure 6. Hh pathway down-regulates Hgf via miR-26a and miR-26b.
(a) Schematic representation of two predicted miR-26a target sites within the Hgf 3′UTR. Vertical lines indicate Watson-Crick pairing. Two nucleotides were mutated in each target site. Numbers indicate positions of nucleotides in the reference sequence (NM_010427). (b) pMir-Report luciferase vectors used to test if miR-26a/b target the Hgf 3′UTR. Luc-Ctrl contains the luciferase gene only. Luc-Hgf-FL contains the luciferase gene with the entire Hgf 3′UTR. Luc-Hgf-Mut contains the luciferase gene with the entire Hgf 3′UTR, with two nucleotides mutated in both sites (a). (c) Vectors in (b) were transfected into HEK293S cells with miR-26a or miR-26b mimics or a control scrambled (Neg) miRNA, and the pRL-SV40 Renilla luciferase vector. The y-axis shows luciferase normalized to renilla activity. Data are mean ± s.e.m. of separate transfections (n = 3). Transfection of miR-26a or miR-26b led to a 45% and 30% decrease in luciferase activity of Luc-Hgf-FL but not Luc-Ctrl or Luc-Hgf-Mut compared to the control. One representative experiment of three is shown. (d) Experimental scheme to test the effect of miR-26a or miR-26b on Hgf levels in primary prostate stromal cells. (e) Transfection of miR26a or miR26b mimics led to a 30% and 31% decrease in Hgf protein secreted into the media, and a 40% and 30% decrease in Hgf transcripts respectively (f). (e,f) n=3 wells per condition, one representative experiment of three shown. (g) Experimental scheme to test the effect of purmorphamine on miR-26a and miR-26b levels in prostate stromal cells. (h) Treatment with purmorphamine led to a 1.2-fold increase in miR-26a levels and a 1.3-fold increase in miR-26b levels compared to control (DMSO). n=3 wells per condition, one representative experiment of three shown. (i) RT-PCR analysis of RNA isolated from EYFP-positive (EYFP+) and EYFP• negative (EYFP−) stromal cells from Gli1CreER/WT; R26EYFP/WT prostates during regeneration (see Fig. 3g). EYFP+ cells express higher levels of miR-26a and miR-26b than EYFP− cells. n=3 technical replicates, one representative experiment of three is shown. (c,e,f,h,i) Data are presented as mean ± s.e.m., significance was calculated by a unpaired Student’s t-test (*: p<0.05, **: p<0.01,***:p<0.001).