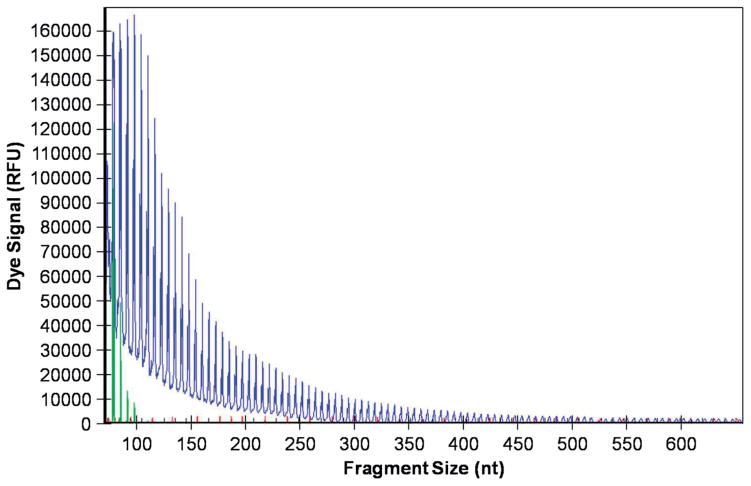
Figure 2.
C9orf72 (G4C2) repeat expansion detection using repeat-primed PCR. Genomic DNA was amplified using a three-primer system and touchdown PCR cycling program as detailed in the methods and previously described (22,23). In brief, the forward primer contains a 5′ fluorescently labeled tag that hybridizes upstream of the (G4C2) repeat expansion, the reverse primer consists of a non-complementary 5′ tail sequence and three (G4C2) repeat units, and the anchor primer corresponds to the anchor sequence of the reverse primer. As shown in the representative electropherogram, the initial peak corresponds to the shortest fragment containing a minimum of three (G4C2) repeats, with each successive peak corresponding to fragments expanding by one repeat unit. A characteristic saw-tooth pattern is generated as signal intensity decreases as fragment size increases. Electropherograms displaying ≥30 repeats is commonly used as the pathological cut-off value (23).