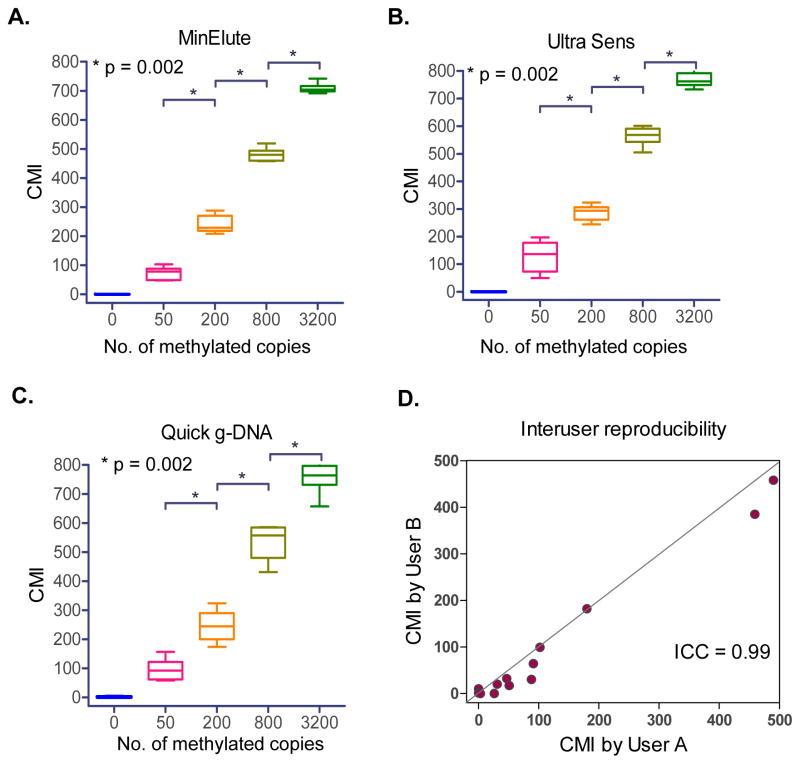
Figure 2. cMethDNA Assay Validation.
A–C) Three serum purification methods (Qiagen MinElute, Qiagen Ultra Sens, and Zymo Research Quick g-DNA) were tested in conjunction with the cMethDNA method in normal serum aliquots spiked with 0–3200 methylated copies, prepared from one master stock. Replicates were purified and multiplexed separately. Box-Whisker plots show the median and full range of CMI (Y-axis) for replicates of each sample (X-axis). Statistical significance (Mann-Whitney test) is indicated by p-values. The % coefficient of variation (CV, a normalized measure of frequency distribution) is shown for each test in Supplementary Table 4. D) Inter-user reproducibility was evaluated for a set of thirteen patient serum samples processed independently by two investigators. User performance was evaluated for the 10-gene panel (Intra-class correlation coefficient = 0.99, 95% CI = 0.96–1.00).