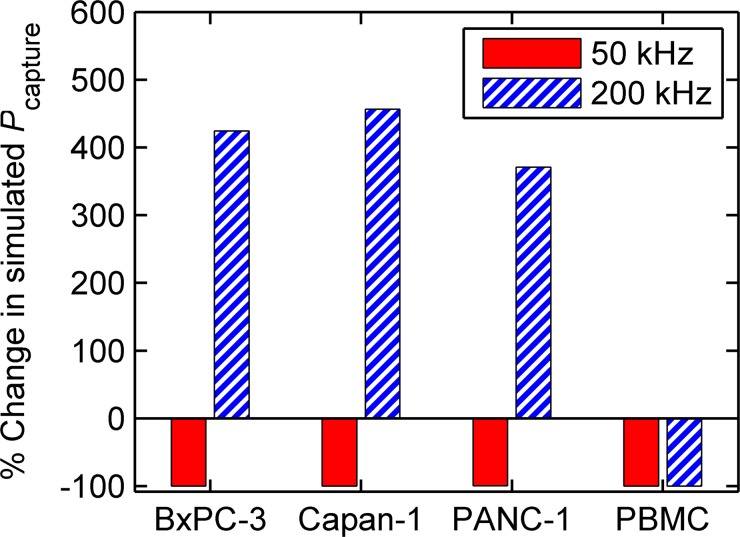
FIG. 5.
DEP significantly effects the capture probability of cells in the obstacle array. Here, the percent change in simulated —i.e., —is shown for pancreatic cancer cells and PBMCs for two independent simulations, one driven at 50 kHz and another at 200 kHz, in an array with Δ = 2 μm. The simulations show that the capture of contaminating PBMCs is reduced by greater than 99% at 200 kHz (from 0.16 to 0.001); simultaneously, the capture of target pancreatic cancer cells is increased by 370%–450% (from 0.03–0.10 to 0.14–0.55, based on cell line). Although the absolute capture probability for pancreatic cancer cells would be greater at a larger offset, the improvement in sample purity (i.e., the large reduction in Pcapture for PBMCs) predicted using DEP at a small offset outweigh a small decrease in capture efficiency, especially for applications such as single-cell genetic analyses.