Figure 2.
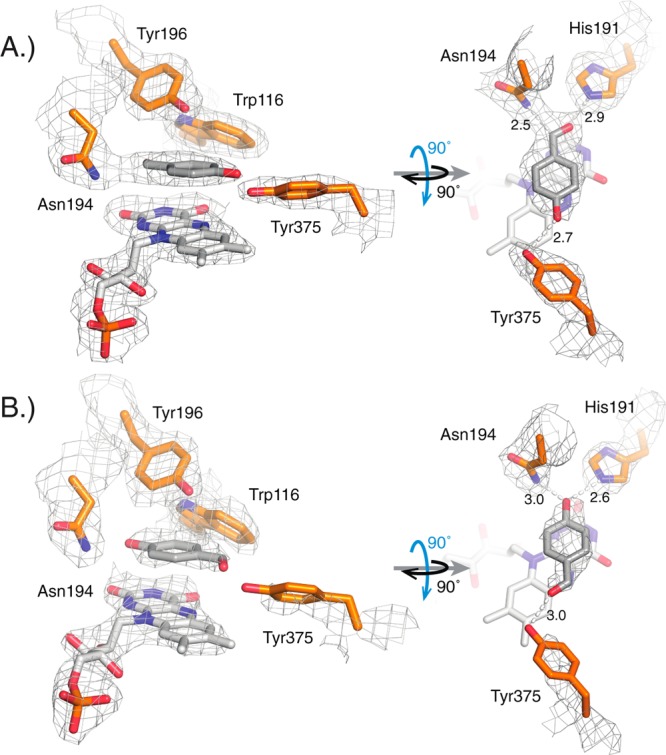
Structural study of the active site of cpOYE303 (PBD: 4RNV) with key amino acid side chains (orange), FMN (white), and substrate analog p-hydroxybenzaldehyde (HBA; gray). (A) In two of the four protein complexes per asymmetric unit, the best fit to the observed electron density orients the aldehyde group of HBA in hydrogen-bonding distance to H191 and N194, whereas in the other two complexes (B), the ligand is flipped with its hydroxyl moiety pointing toward H191 and N194, as seen for OYE1.33 The gray mesh represents the 2mFo-DFc map contoured at the 1.0 σ level for key protein side chains, FMN and HBA. Hydrogen bonding distances (in angstroms) are indicated.
