Figure 2.
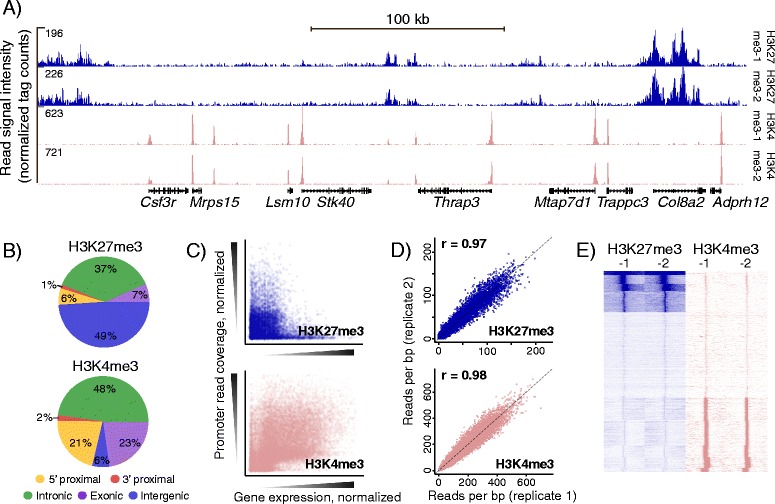
Validation of small-scale ChIP-seq histone mark maps. (A) Two H3K27me3 and H3K4me3 profiles and Refseq gene positions across a 400 kb region on chromosome 4. (B) Genomic distribution of all mapped reads, rounded numbers. (C) Scatterplots of promoter histone methylation and mRNA expression in GMP-phenotype blast cells. Each data point represents a single gene. Quantile normalized. (D) Quantitative comparisons of promoter read coverage (reads per kilobase) from two independent biological replicates of each histone modification using 1-kb TSS centered bins. Quantile normalized, 1/1 diagonal indicated by dashed line (r = pearson correlation coefficient, calculated on normalized data, 1 outlier pre-filtered). (E) SeqMiner cluster heatmaps showing signal intensities for both replicates of H3K27me3 and H3K4me3 in a 20000 bp region centered on Refseq TSS positions.
