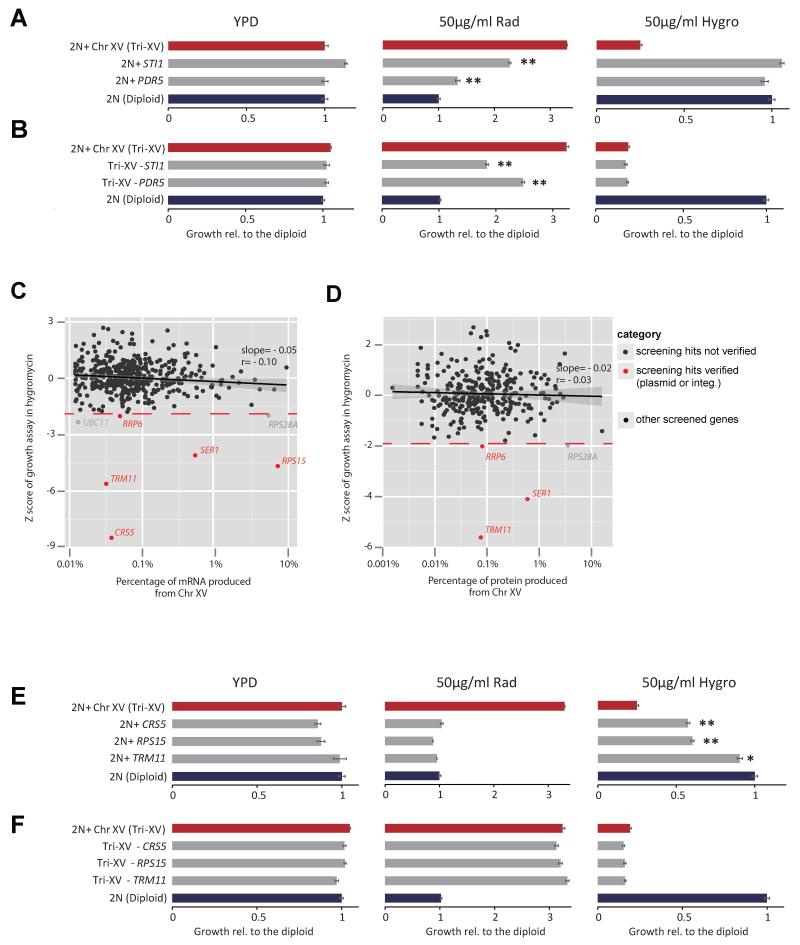
Figure 4. Different sets of genes on Chr XV cause radicicol sensitivity or hygromycin B sensitivity when increased in copy number.
(A-B) Copy number gain (+, by genomic integration) or loss (−, by genomic deletion) assays showing that increased copy numbers of STI1 and PDR5, which are both critical for radicicol resistance, do not contribute to the hygromycin B hypersensitivity. Relative growth compared to the diploid control is reported in bar plots with the SEM derived from 3 replicates. Asterisks denote significant difference from the corresponding control (the diploid or Chr XV trisomy (Tri-XV)) (*, p< 0.05; **, p<0.01; two-tail t-test). (C-D) Each of 453 genes located on Chr XV was transformed into a diploid strain,and Z scores denoting the deviation of growth of each strain from the population average in the presence of 35 μg/ml hygromycin B were plotted against mRNA (C), using RNAseq data, or protein expression abundance (D) (Ghaemmaghami et al., 2003), of each tested gene in the euploid S288c background. Note that protein abundance data were not retrieved for 30% genes (including CRS5 and RPS15). The grey area shows the 95% confidence interval for the linear fitting. 35 μg/ml hygromycin B produced similar growth inhibition to euploid control in SC –ura media compared to euploid control in YPD media. (E) Growth assays showing that copy-number increase (by genomic integration) of 3 genes (CRS5, RPS15, TRM11) on Chr XV were individually sufficient in a diploid euploid context to reproduce enhanced sensitivity to hygromycin B, but not radicicol resistance, contrasting copy number increase for STI1 and PDR5 as shown in A. (F) Growth assays showing that single-copy deletion of none of the 3 genes (CRS5, RPS15, TRM11) alone could rescue Chr XV trisomy from hygromycin B hyper-sensitivity.
See also Figure S5.