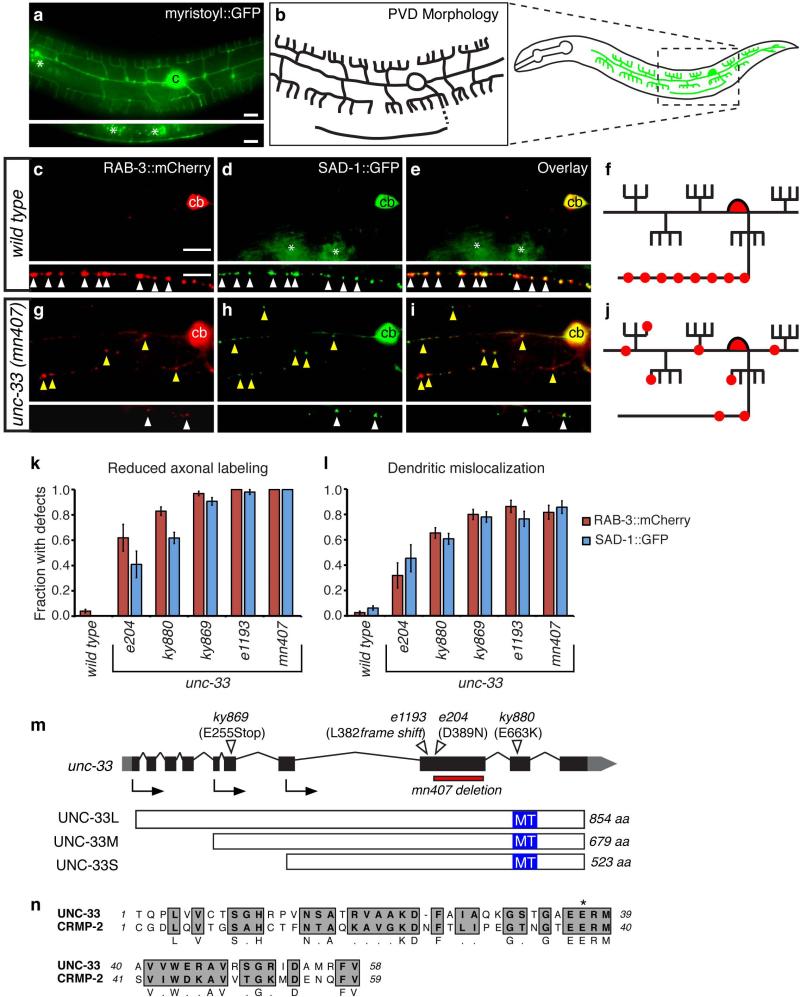
Figure 1. unc-33 mutants mislocalize presynaptic proteins to dendrites.
(a,b) PVD neuron morphology. (a) des-2::myristoyl::GFP marker in an L4 animal (b) Diagrams of PVD morphology. The unbranched ventral process is the axon.
(c-j) Representative images of RAB-3::mCherry and SAD-1::GFP in PVD neurons of wild type (c-f) and unc-33(mn407) (g-j) animals. For each set of fluorescence micrographs, the top panel is the maximum intensity projection of dendritic focal planes and the bottom panel is the maximum intensity projection of axonal focal planes. White and yellow arrowheads indicate axonal and dendritic puncta, respectively; ‘cb’ labels the PVD cell body, and asterisks mark gut autofluorescence. Anterior is at left and dorsal is up in all panels. Scale bars, 10 μm.
(k,l) Quantification of axonal localization defects (k) and dendritic mislocalization defects (l) of RAB-3::mCherry and SAD-1::GFP (n>30 animals/genotype). The fraction of animals with qualitative defects is shown. Error bars indicate standard error of proportion (s.e.p.). Alternative quantification of fluorescence intensity per animal is provided in Supplementary Fig. 1.
(m) unc-33 gene structure showing exons (black boxes), introns (lines), and untranslated regions (gray boxes), and lesions in unc-33 alleles. Arrows denote the positions of start codons for alternative unc-33 transcripts. Three UNC-33 protein isoforms (long, medium and short) are depicted with predicted microtubule-assembling domains in blue. (n) Predicted microtubule-assembling domain of UNC-33, with rat CRMP-2. Gray regions highlight conserved residues; asterisk denotes the conserved glutamate mutated to lysine in unc-33(ky880).