Figure 6. Human Pain-Nociceptive and inflammatory PCR array.
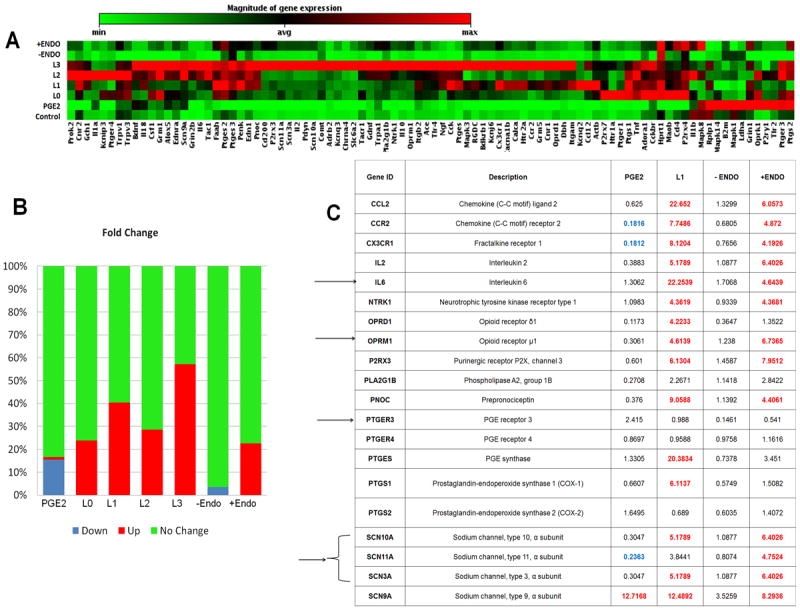
Human Pain-Nociceptive and Inflammatory PCR array on Ishikawa endometrial cell lines that were treated with either PGE2 (50ng/ml) or the different forms of oxidatively modified LDLs (25μg/ml), or PF (100μl) from patients with endometriosis (+ Endo) or control women (-Endo) for 48 hours. (A) Clustergram of gene expression in cells treated with PGE2, oxidatively modified LDLs and peritoneal fluid. The array includes 84 inflammatory and nociceptive genes as well as housekeeping genes and quality control standards. (B) Represents the percent of genes that were classified as up-regulated, down-regulated, or no change compared to the control treatment based on a 4-fold cutoff of the fold change. (C) Represents notable genes based on fold change. Potential genes involved in the oxidatively-modified LDL mediated nociceptive pathway are highlighted with an arrow. While PGE2 and PF from women without endometriosis (-Endo) had very little change in gene expression, oxidatively-modified LDLs and endometriotic fluid from patients with endometriosis (+Endo) showed similar trends in induction of several nociceptive and inflammatory genes. [The supplementary Table 1 lists all the genes measured in the pain array]. Statistical analysis (T-test) was performed using the algorithm provided by the manufacturer (Qiagen).
