Figure 2. Sample retrieval of average change in information content (Δ R i) with splicing mutation calculator (SMC) for published mutations.
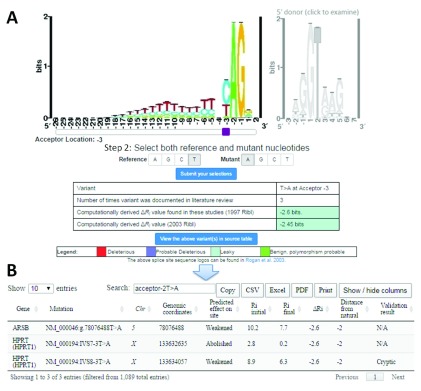
A) Example mutation input for SMC (T>A at the 3 rd intronic position of natural acceptor). The type of splice site is selected by clicking on the corresponding sequence logo (acceptor [left] or donor [right]). The purple slider bar appearing below the logo is used to select the position of the mutation. The reference and mutant nucleotides are then designated, and the variant is submitted to the software (‘Submit your selection’). SMC outputs a table indicating the user input, the number of instances in the literature where this substitution has been analyzed using IT, and the computed Δ R i values (in bits) using both the old (1992; top) and new (2003; bottom) ribls. The cell color for Δ R i values indicates the predicted severity of the inputted variant according to defined thresholds 22, 168. B) Tabular output detailing each instance of the selected mutation from the source table. The user may view, in a separate window, extensive details of all variants referred to in SMC output ( Supplementary Table 10).
