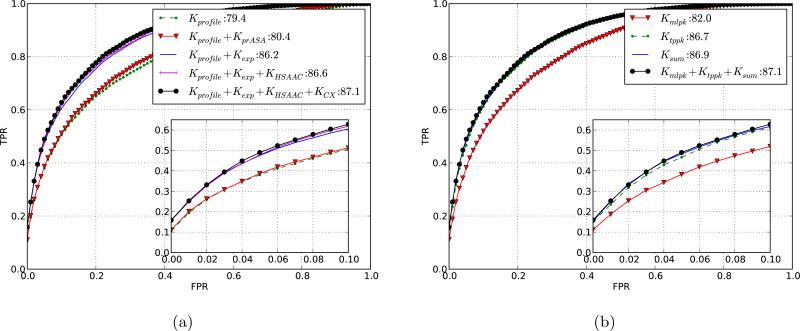
Figure 3.
Selecting the optimal sequence/structure representation by comparing ROC curves for different kernel designs. Shown are the averaged ROC curves computed using 5-fold cross-validation over complexes in DBD 3.0. The inset shows the true positive rate (TPR) vs. false positive rate (FPR) for up to first 10 % false positives. The legend shows the AUC scores for the different kernels used. (a) Results for different residue kernels Kr using the pairwise kernel Kpw = Kmlpk + Ktppk + Ksum. The curves illustrate the increase in performance as additional structural information is added to the sequence-based kernel. Recall that Kprofile is the PSI-BLAST profile kernel; KprASA uses predicted rASA; Kexp is the residue exposure kernel; KHSAAC is the half-sphere exposure kernel; KCX uses protrusion-index features. (b) Results for different pairwise kernels Kpw with residue kernel Kr = Kprofile + Kexp + KHSAAC + KCX.