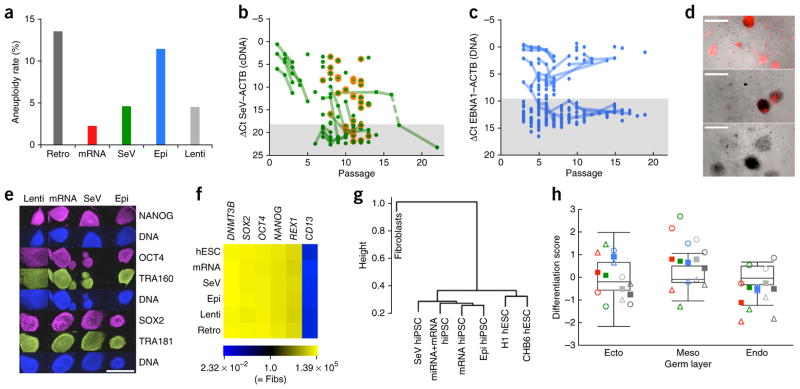
Figure 2.
Comparison of the genetic integrity of hiPSCs derived by different reprogramming methods. (a) Aneuploidy rates of low-passage (P < 30) hiPSC lines derived by different reprogramming methods. The data include only standard reprogramming methods and the results from the lowest available passage for each hiPSC line. All observed abnormal karyotypes are listed in Supplementary Figure 1a. Donor age did not contribute to the increased rate of aneuploidy among Epi hiPSCs (data not shown). n = 192 (Retro), 44 (mRNA), 151 (SeV), 61 (Epi), 22 (Lenti). (b) Quantification of SeV (Cytotune) RNA in SeV hiPSC lines at different passages by TaqMan RT-QPCR analysis. Ct values for ACTB were subtracted from those for SeV. Lines connect data points that represent the same stem cell line over time. A 5-day heat treatment (39 °C) was performed between collections of the samples connected by the dashed line. The shaded area marks the detection limit (3 s.d. removed from the mean of the SeV-negative controls (n = 10)). Orange circles highlight erythroblast-derived SeV hiPSC samples showing a slightly delayed loss of SeV RNA. Cytotune2 SeV RNA loss kinetics are shown in Supplementary Figure 2a. (c) Quantification of EBNA1 DNA in Epi hiPSC lines at different passages by TaqMan QPCR analysis. Ct values for RNAseP (single-copy control gene locus) were subtracted from those for EBNA1. Lines connect data points that represent the same stem cell line over time. The shaded area marks the detection limit (3 s.d. removed from the mean of the EBNA1-negative controls (n = 41)). See also Supplementary Figure 2b. (d) Epi hiPSCs (passage 3) generated with a modified OCT4-p53 plasmid containing a H2B-mKO2 cassette for fluorescent labeling of hiPSCs that have retained this plasmid (shown are examples of lines containing varying amounts of H2B-mKO2; some dead show auto-fluorescence). Scale bars, 1,000 μm. (e) Immunofluorescence analysis of hiPSC lines representing the indicated reprogramming methods. Scale bar, 1,000 μm. (f) Quantitative RT-PCR analysis (passage 10–15) of fibroblast and pluripotency marker gene expression in parental fibroblasts and hiPSCs, derived by the indicated methods. Heat map representation of the average fold-induction and fold-repression (compared to fibroblasts) of GAPDH-normalized expression levels of three hiPSC lines per method (three technical replicates per line). Additional genes are shown in Supplementary Figure 3. (g) Hierarchical cluster analysis of CpG genomic DNA methylation levels of partially methylated domains in method-specific hiPSC (passage 11–21) and control cells (Supplementary Fig. 4). (h) Comparison of the scorecard differentiation propensity of method-specific hiPSC lines (passage 15–27). Boxplots show the distribution of differentiation propensities for the pluripotent reference set (hESCs); circles = BJ-derived hiPSCs, triangles = PS1-derived hiPSCs; solid squares = method-specific score averages; for each germ layer, method-specific hiPSC scorecard data are shown (from left to right) for mRNA (red), SeV (green), Epi (blue), Lenti (light gray) and Retro hiPSCs (dark gray). None of the pairwise scorecard-score comparisons (mRNA vs. Epi, mRNA vs. SeV, Epi vs. SeV) reached statistical significance (Student’s t-test, P > 0.5).