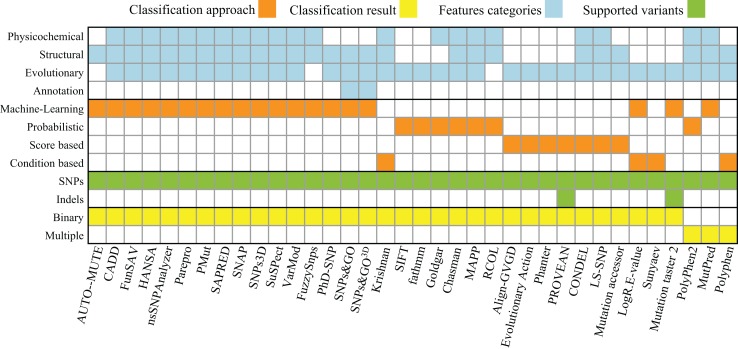
Figure 2.
Summary of properties and approaches for software listed in Table 1. The approaches found fall into four different categories: Machine-Learning, Probabilistic, Score (calculating a summarizing score of a set of hand-picked statistics), and Rule (using a set of empirically derived rules). These approaches provide one of two types of classifications each: a binary classification (e.g., neutral or deleterious) or a multi-classification (e.g., benign, neutral, and deleterious). The features used by those approaches can be computed based on properties of the following five categories: (i) physicochemical properties (e.g., solvent accessibility, polarity, charge, disorder, and Grantham), (ii) structural information about the primary, secondary, and tertiary structure of a protein (e.g., α-helices, β-sheets, and coil), (iii) evolutionary properties (multiple sequence alignments, position-specific scoring matrices, and Hidden Markov models), and (iv) genome annotation (GO terms or other protein function annotations). The supported variants were determined either by accessing the tools’ websites or by the description of the approach itself.