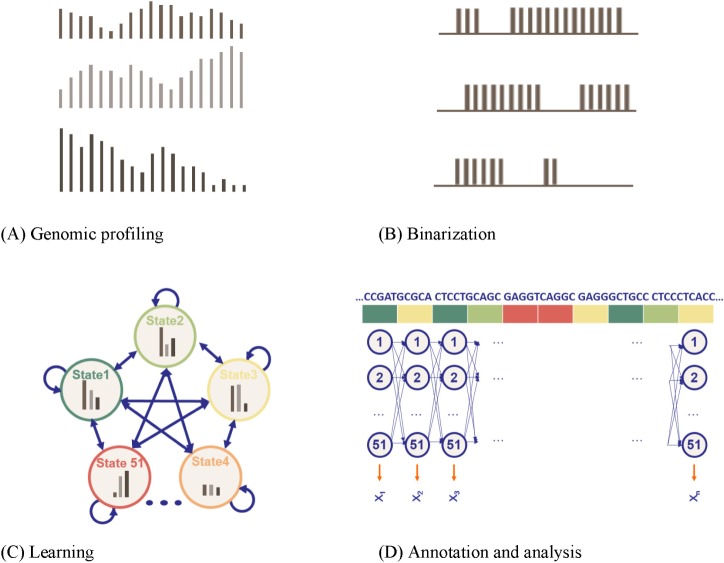
Fig. 5.
A multivariate, multi-state hidden Markov model (HMM) [19]. (A) Genomic profiling. To apply the model, the genome was divided into 200-base-pair nonoverlapping intervals, within which each of the count of 41 marks that mapped to the interval was annotated. (B) Binarization. For each 200-bp interval, the input ChIP-Seq sequence tag count is processed into a binary presence/absence call. (C) Learning. Each model was scored based on the log-likelihood of the model minus a penalization on the model complexity, determined by the Bayesian Information Criterion (BIC). (D) Annotation and analysis. The vector of 41 numerical values was assigned, each representing the result of a different biochemical assay, and each of the 200-base-pair intervals was assigned to its most likely state under the model (picture adapted from the cs262 class slides by S. Batzoglou with permission).