Fig. 1.
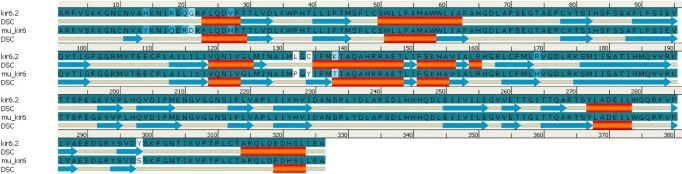
Comparison of predicted secondary structures in wild-type (kir6.2) and mutant-type (mu_kir6.2) model. Alignment done for amino acids 33-358 of kir6.2 models. Secondary structural element (DSC) helix is represented by red cylinder, and beta sheets are represented by blue arrows in the picture.
