Figure 5.
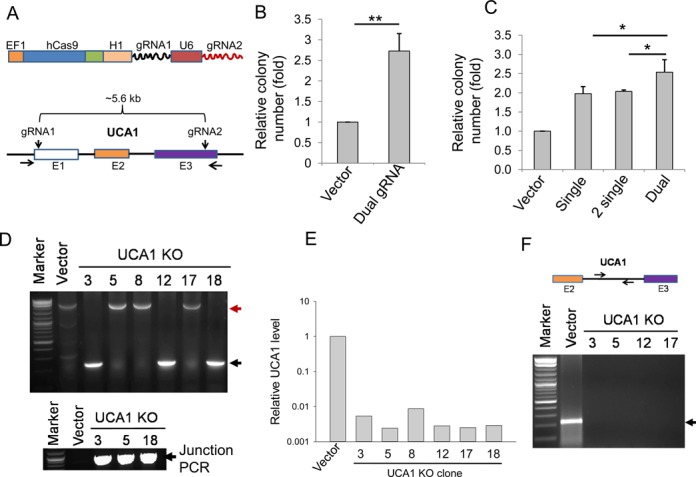
Targeting UCA1 with a dual gRNA approach. (A) Strategy for targeting UCA1 with dual gRNAs. (B) Colony formation for vector and UCA1 KO after puromycin selection. (C) The dual gRNA approach increases colony number, as compared to single or two individual gRNAs in combination. (D) Detection of UCA1 deletion by genomic PCR using primers UCA1-outside-5.5 and UCA1-outside-3.5. The red arrow (2.8 kb) indicates a PCR product corresponding to a replacement and the black arrow indicates a deletion (227 bp). Clones #3, #12 and #18 are expected to have a 2.8-kb fragment, but missing here possibly because a deletion band is much smaller, and thus it is more favorably amplified. Indeed, junction PCR with primers UCA1-Recom-5.1 and EF1–seq-3.2 revealed that clones #3, #5 and #12 all carried the HR fragment (bottom). (E) Detection of UCA1 expression in UCA1 KO clones by qRT-PCR. P value <0.01 for all six clones, as compared to vector control. (F) No PCR product (bottom) except for vector control (220 bp) was detected in UCA1 KO clones using primers derived from the region as indicated on top (UCA1-intron-5.1 and UCA1-intron-3.1). Values in (B) and (C) are means of ± SE (n = 3). *P < 0.05 and **P < 0.01.
