Figure 2.
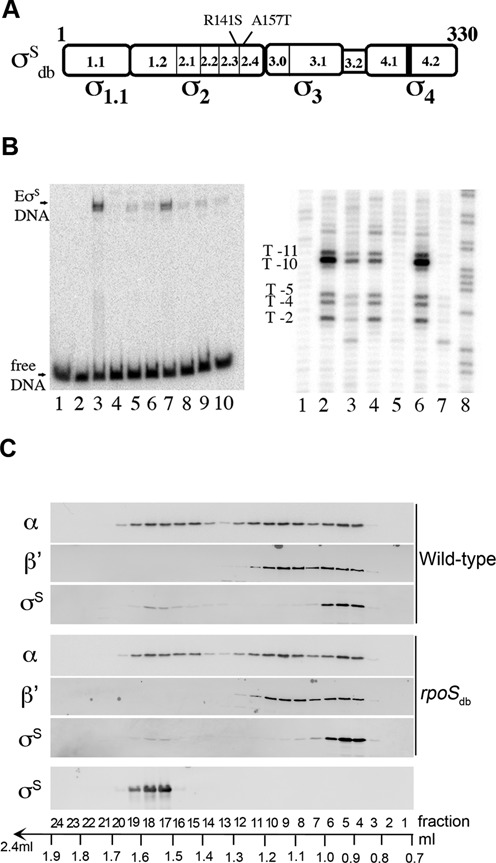
σSdb is impaired in DNA promoter binding but not in RNAP core binding. (A) Schematic representation of the four regions of the σS protein (17,51) and location of the amino-acid substitutions in σSdb in regions involved in binding and melting the -10 promoter element (54,55). (B) The σS variants containing one or both substitutions are deficient in DNA promoter binding. When bound to core RNA polymerase (E), σS makes sequence-specific promoter contacts and plays a crucial role in DNA melting (54–56). Left panel: electrophoretic mobility shift assay indicating binding of EσS, but not EσSdb, to the katN promoter region. The [32P]-labeled katN promoter fragment was incubated for 20 min at 37°C with buffer (lane 1), E (lane 2) or holoenzymes reconstituted with His6-σS wild-type (lanes 3 and 7), His6-σSR141S (lanes 4 and 8), His6-σSA157T (lanes 5 and 9) and His6-σSdb (lanes 6 and 10) before heparin challenge. The E:σS ratios were 1:33 (lanes 3–6) and 1:4 (lanes 7–10). Right panel: KMnO4 probing of the EσS holoenzymes (E:σS of 1:10) on the 5′-[32P]-labeled template strand katN fragment. KMnO4 preferentially oxidizes exposed unstacked thymines of RNAP–promoter complexes and gives rise to marked KMnO4 reactivity at the katN promoter as previously reported (49). The positions of the reactive Ts with respect to the transcription start are indicated (T-11, T-10, T-5, T-4 and T-2). Lane 1: control with no protein; lanes 2 and 6: E His6-σS wild-type; lane 3: E His6-σSR141S; lane 4: E His6-σSA157T; lane 5: E His6-σSdb; lane 7: E only. Lane 8 is a G+A reaction performed on the same DNA fragment according to Maxam and Gilbert (58). (C) Distribution of σS between free and holoenzyme forms in the wild-type and rpoSdb strains. Whole cell lysates from wild-type strain VF7969 and rpoSdb mutant VF9849 were fractionated by size exclusion chromatography and the relative concentration of RNAP subunits was subsequently analyzed in the fractions by immunoblot using monoclonal antibodies against the β’ and α subunits of RNA polymerase and a polyclonal anti-σS antibody. Purified σS was used to pinpoint fractions containing free σS. The percentage of total σS in fractions corresponding to free σS was very low (and appeared slightly lower in rpoSdb than in wild-type) suggesting that most σS molecules are associated with RNAP in stationary phase. Two independent experiments were performed with similar results (the elution profiles were similar for the wild-type strain and the rpoSdb mutant, and the percentages of total σS in fractions 4–8 (bound σS) were 69 and 83% for the wild-type strain and 83 and 93% for the rpoSdb mutant, calculated using the IMAGEJ software).
