Figure 1.
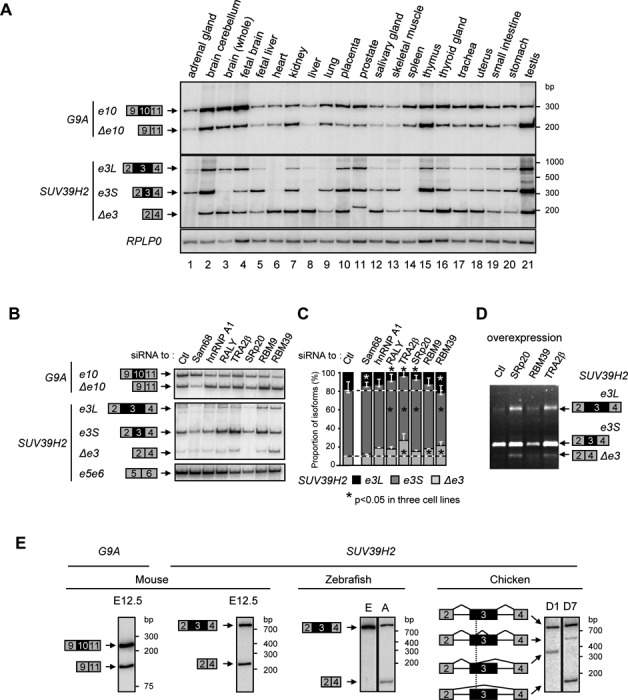
Alternative splicing regulates the expression of G9A and SUV39H2. (A) Analysis in 21 human tissues of inclusion of G9A exon 10 (e10, exon 10 included; Δe10, exon 10 skipped) and SUV39H2 exon 3 (e3L, exon 3 fully included; e3S, exon 3 partially included; and Δe3, exon 3 skipped). Semi-quantitative RT-PCR was performed on total RNA using radiolabeled primers. PCR products are indicated on the left, and level of RPLP0 transcripts were used as control. (B) Regulation of G9A exon 10 and SUV39H2 exon 3 by splicing factors. The alternative inclusion of G9A exon 10 and SUV39H2 exon 3 was assessed in HeLa cells by siRNA-mediated depletion of indicated factors. Various spliced isoforms were amplified as in the panel A, while SUV39H2 exons 5 and 6 were amplified as control (bottom). (C) Ratio of SUV39H2 spliced isoforms in HeLa cells were quantified by capillary electrophoresis. Data display an average of two experiments, and stars indicate significant changes of isoforms in three cell lines (see also Supplementary Figure S1I) estimated by Wilcoxon-ranked test (P-value < 0.05). (D) Overexpression in HeLa cells of indicated splicing factors affects SUV39H2 exon 3 splicing. SUV39H2 isoforms were amplified by RT-PCR and analyzed in agarose gel stained with ethidium bromide. Splicing patterns displayed a representative example of three experiments. (E) Analysis in different organisms of alternative splicing decisions for orthologous G9A exon 10 and SUV39H2 exon 3. Semi-quantitative RT-PCR was performed with radiolabeled primers and total RNA extracted from: E12.5 mouse embryos, Zebrafish embryos (E) and adult (A), chicken embryos at day 1 (D1) and day 7 (D7). For chicken, the various alternative splicing events of SUV39H2 exon 3 are indicated with sharp lines. Inclusion of orthologous G9A exon 10 was only tested in mouse as the structure of its gene is not conserved in other species.
