Figure 2.
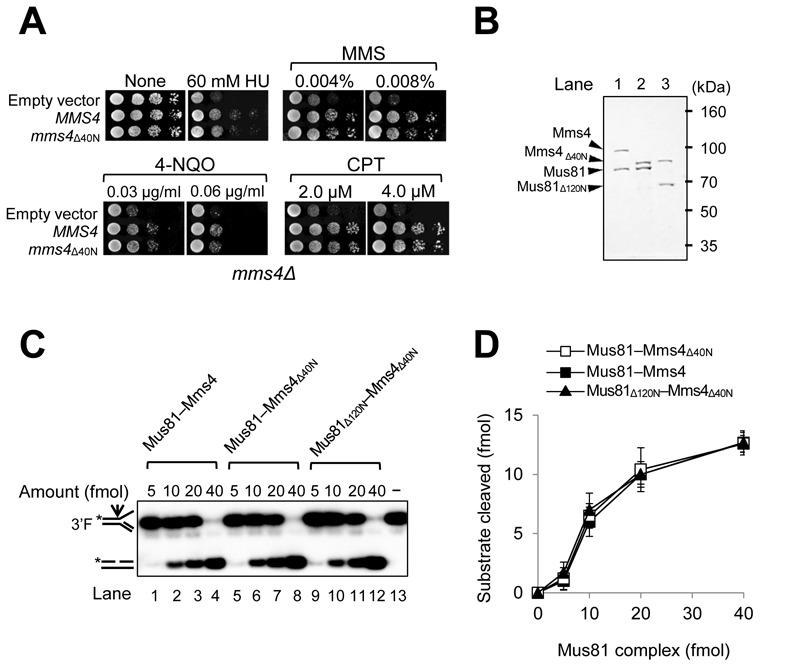
The N-terminal 120 aa region of Mus81 is dispensable for the catalytic endonuclease activities of Mus81 complexes. (A) Complementation activity of mms4Δ40N to rescue the drug sensitivity of mms4Δ strain. MMS4 or mms4Δ40N under the control of its native promoter in the pRS314 vector was transformed into HY1728 strain (mms4Δ). The transformants were grown until saturation in liquid media and then spotted onto plates with indicated amounts of DNA damaging agents. The plates were incubated at 30°C for 3 days. HU, hydroxyurea; MMS, methyl methanesulfonate; 4-NQO, 4-nitroquinoline oxide; CPT, camptothecin. (B) Purification of recombinant Mus81–Mms4, Mus81–Mms4Δ40N and Mus81Δ120N–Mms4Δ40N complexes. SDS-PAGE (10%) analysis of purified Mus81 complexes, followed by Coomassie blue staining. The sizes of molecular mass marker are indicated in kDa. (C) Comparison of endonuclease activities of three recombinant Mus81 complexes from panel (B). Reactions were carried out in standard reaction mixtures containing 15 fmol of 3′F substrate and increasing amounts (5, 10, 20 and 40 fmol) of Mus81 complexes. Reactions were incubated at 30°C for 30 min and terminated by the addition of 0.2% SDS, 10-μg proteinase K, followed by incubation at 37°C for 15 min. The products were subjected to a 10% PAGE in 0.5X TBE at 150 V and the gels were dried and autoradiographed. The structures of 3′F substrate and the cleavage product are as illustrated in the figure. The arrow on the substrate denotes the site of cleavage. Asterisks indicate the position of 32P-label at the 5′ DNA ends. (D) The amount (fmol) of cleavage products formed by the endonuclease activity of Mus81 complexes on 3′F substrate (in panel (C)) was plotted against the amount (fmol) of Mus81 complexes used. The graph with error bars indicated was obtained from four independent experiments. The error bars represent the standard deviation from the mean of four independent experiments.
