Figure 7.
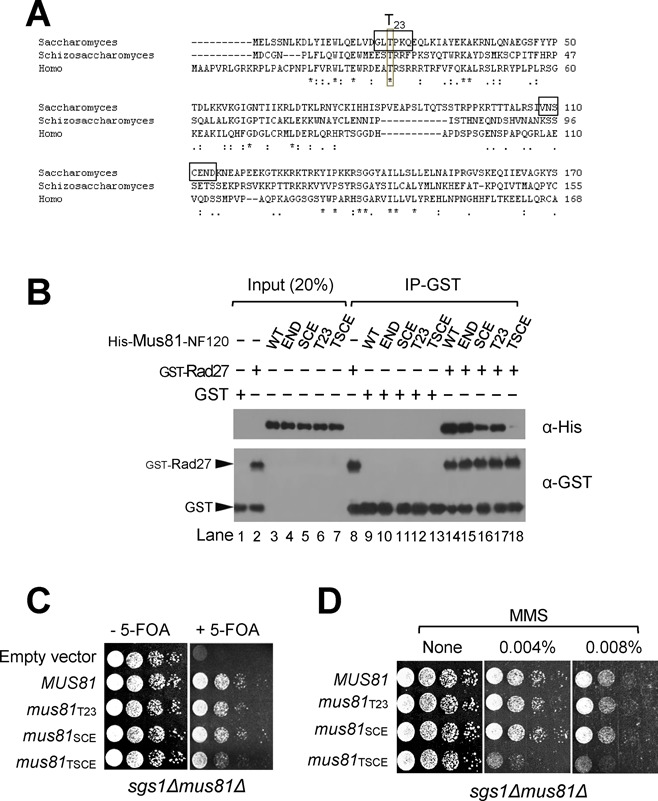
Both N21–26 and N108–114 motifs are important for the cellular function of Mus81–Mms4 complex in the absence of Sgs1. (A) Alignment of the N-termini of Mus81 subunits from Saccharomyces cerevisae, S. pombe and H. sapiens. The N21–26 and N108–114 motifs in yeast Mus81 N-terminus are indicated by opened boxes. (B) GST pull-down assays with GST-Rad27 and derivatives of Mus81-NF120 (WT) including Mus81-NF120END (END), Mus81-NF120SCE (SCE), Mus81-NF120T23 (T23) and Mus81-NF120TSCE (TSCE). All aa residues of interest (T23, S110, C111, E112, N113 and D114) were changed into alanine. The GST pull-down assays were carried out as described in the Materials and Methods section. (C) The complementation of sgs1Δmus81Δ synthetic lethality by mus81 mutant alleles that contained either T23→A or S110C111E112→AAA or both substitutions. NJY1777 (sgs1Δmus81Δ + pJM500-URA3-SGS1) strain was transformed with an empty vector or vectors containing MUS81 and mus81 mutant alleles driven by ADH1 promoter. The transformants were grown until saturation in liquid media and then spotted onto plates with or without 5-FOA. (D) The MMS sensitivity of mus81 mutant alleles was examined in sgs1Δ background. The colonies that formed on the 5-FOA plates from (panel (C)) were inoculated in liquid media and then spotted onto plates without or with indicated amounts of MMS. The plates were incubated at 30°C for 4 days.
