Figure 5.
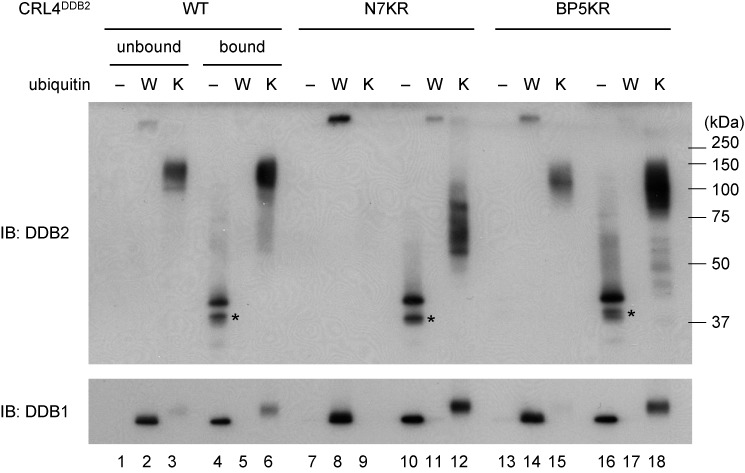
Poly-ubiquitination, but not mono-ubiquitination, abrogates damaged DNA-binding activity of DDB2 regardless of modification sites. The CRL4 E3 ligase complex containing the indicated DDB2 (WT, N7KR or BP5KR) was tested in in vitro ubiquitination assays in the presence of paramagnetic beads bearing DNA containing 6-4PPs. Reactions were performed in the absence of ubiquitin (−), or in the presence of wild-type ubiquitin (W) or K-less ubiquitin (K). Proteins bound or unbound to the DNA beads were separated and subjected to immunoblot analyses. Asterisks indicate degradation products of DDB2 generated during the incubations. DDB1 exhibits a slight band shift only in the reactions containing K-less ubiquitin (lanes 6, 12 and 18), suggesting that DDB1 is not targeted by CUL4 as long as conjugation sites are available on more efficient substrates, such as DDB2 and ubiquitin.
