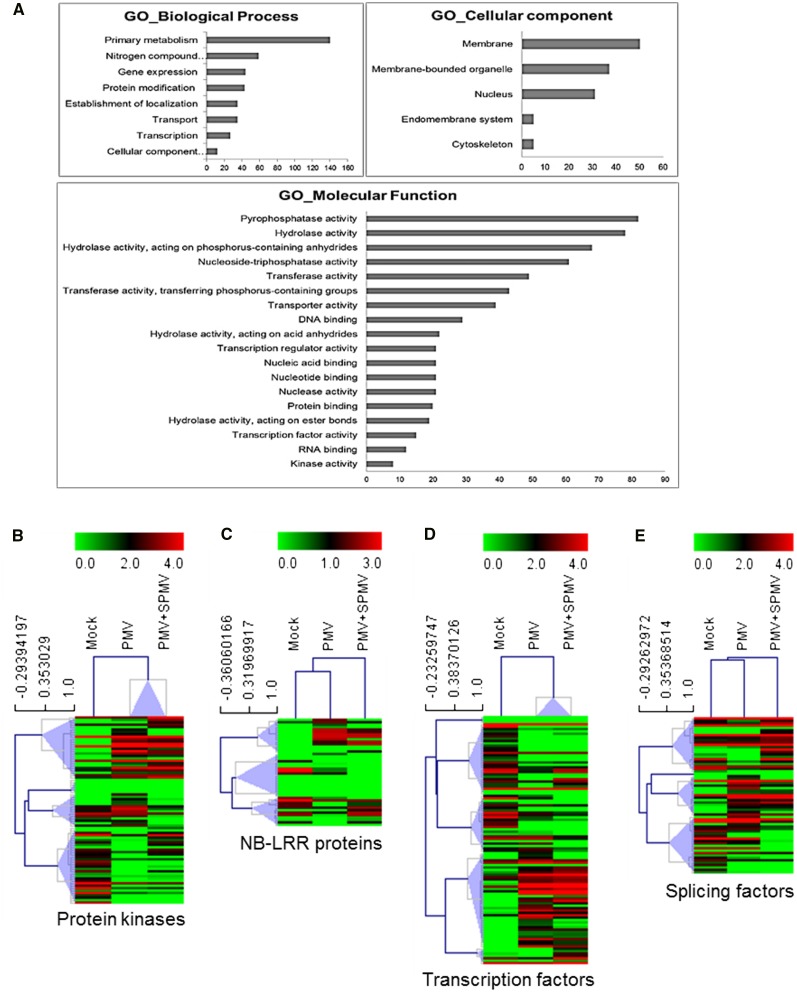
Figure 6.
Virus-Modulated Differentially Spliced Gene Clusters in B. distachyon.
(A) GO biological processes, cellular components, and molecular functions enriched (FDR < 0.05) among differentially spliced genes in PMV and PMV+SPMV infection compared with mock. The x axis indicates the number of genes corresponding to the enriched GO term.
(B) to (E) Hierarchical clustering of expression profiles of the AS transcripts encoding putative protein kinases (B), NB-LRR proteins (C), transcription factors (D), and splicing factors (E). Average linkage method and Pearson correlation metric was used for the clustering. Node heights of the gene clusters are displayed above the gene trees. Colors represent log2 FPKM expression values of the corresponding alternatively spliced transcripts.