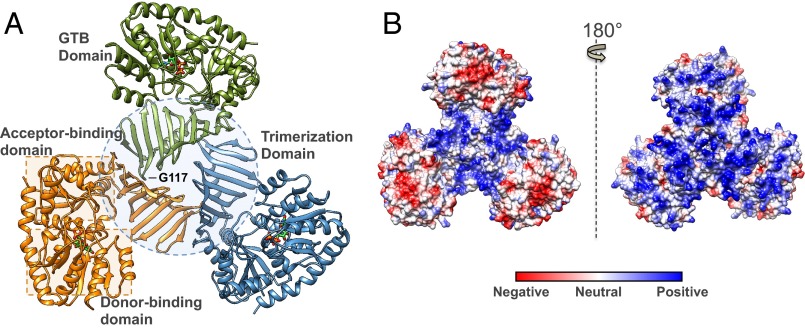
Fig. 1.
Structural features of the S. aureus TarM trimer. (A) Ribbon representation of trimeric TarM in complex with UDP (displayed in stick form and colored according to heteroatom type). The separate protomers are indicated by color, and the trimerization domain and substrate donor/acceptor site Rossmann-like domains are demarcated by a dotted circle and dotted squares, respectively. The G117 residue whose mutation to arginine resulted in the monomeric protein is also marked, showing its remote position from the GT active site. (B) Electrostatic surface representation of the TarM trimer. The molecule on the left is displayed in the same orientation as A, and the molecule on the right upon 180° rotation. The trimer possesses prominent positively charged grooves along its surface, which are localized in the central trimerization site of the oligomer in the left view, and along the entire surface on the right.