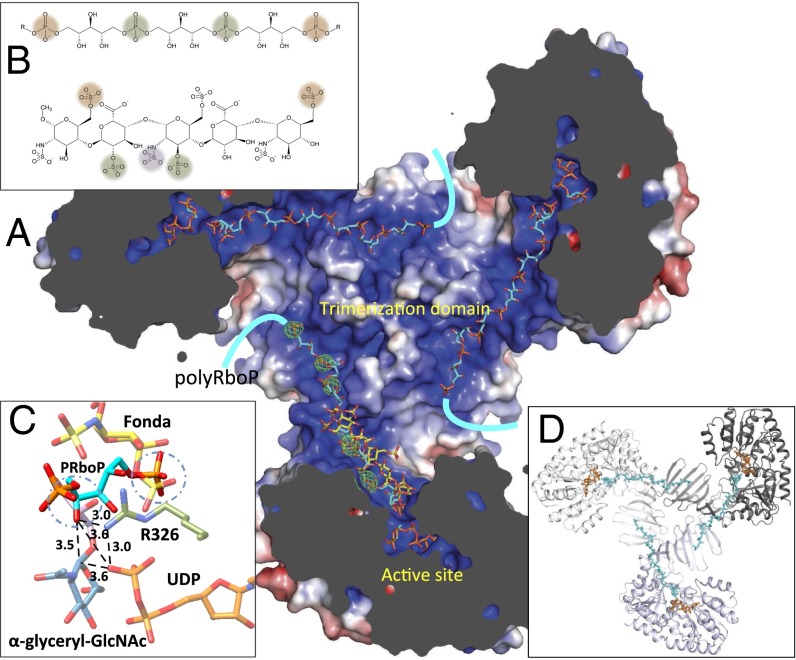
Fig. 4.
Binding of modeled polyRboP to TarM. (A) Model illustrating the proposed binding of an extended polyRboP polymer (cyan) to the positively charged binding groove that extends from the active site along the acceptor binding domain and across to the trimerization domain of a neighboring protomer. Location of ordered sulfate residues in the apo structure are shown in green (Fo-Fc difference map contoured at 4 σ) and the bound fondaparinux (shown for comparison) in yellow. (B) Comparison of polyRboP (Top) and fondaparinux (Bottom) chemical structures. SO4/PO4 groups of respective fondaparinux and modeled polyRboP that overlap with ordered sulfates are displayed in green, and groups that overlap between the two molecules (but not the ordered sulfates) are shown in orange. A nonoverlapping fondaparinux SO4 group that interacts with the electropositive groove in the acceptor binding domain is displayed in purple. (C) Model of the binding of a PRboP unit (cyan) to the active site of TarM. The model places the PRboP C4 hydroxyl (circled) 3.5 Å away from the anomeric carbon (indicated with dashed line), and 3.6 Å from the leaving group phosphate. The PRboP unit is predicted to bind in a similar manner to fondaparinux (yellow), sharing the same phosphate binding pocket as the 6-O-sulfate group of the terminal saccharide of fondaparinux (circled), with the other phosphate coordinated by R326 (green) similar to the fondaparinux sulfoamino group. (D) Same TarM trimer view shown in A but with surface removed to highlight path of polyRboP binding (cyan) across the trimerization interface.