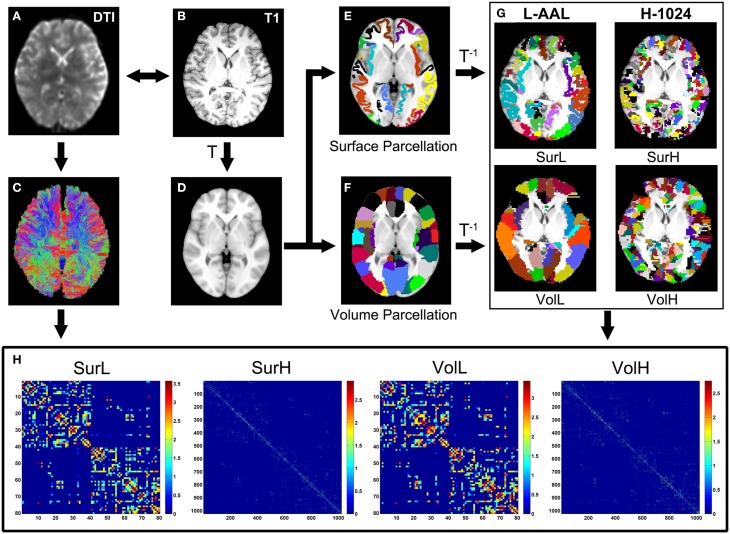
Figure 1.
The flowchart of the construction of four WM networks under two parcellation methods and two resolutions. (1) The b = 0 image (A) and the individual T1-weighted image (B) were coregistered through a linear transformation. (2) The T1 images were then nonlinearly normalized to the ICBM152 T1 template (D) in the MNI space. (3) Each vertex on the average cortical surface of 150 normal brains was assigned with the value of the label in the volumetric AAL (F) to generate an atlas of surface parcellation (E). (4) The inverse transformations were used to warp the AAL atlas to the native diffusion space. (5) Both surface and volume atlases were subdivided into 1024 regions with equal size to define a high resolution nodal scale. (6) The reconstruction of all WM fibers in the brain was performed using deterministic tractography using the Diffusion Toolkit (C). (7) The weighted networks of each subject were created by computing the number of streamlines that connected each pair of brain regions. Both low- (L-AAL) and high-resolution (H-1024) WM networks based on different parcellation approaches (surface and volume) were constructed for each subject (H), which are represented by the abbreviations of SurL, SurH, VolL, and VolH, respectively.