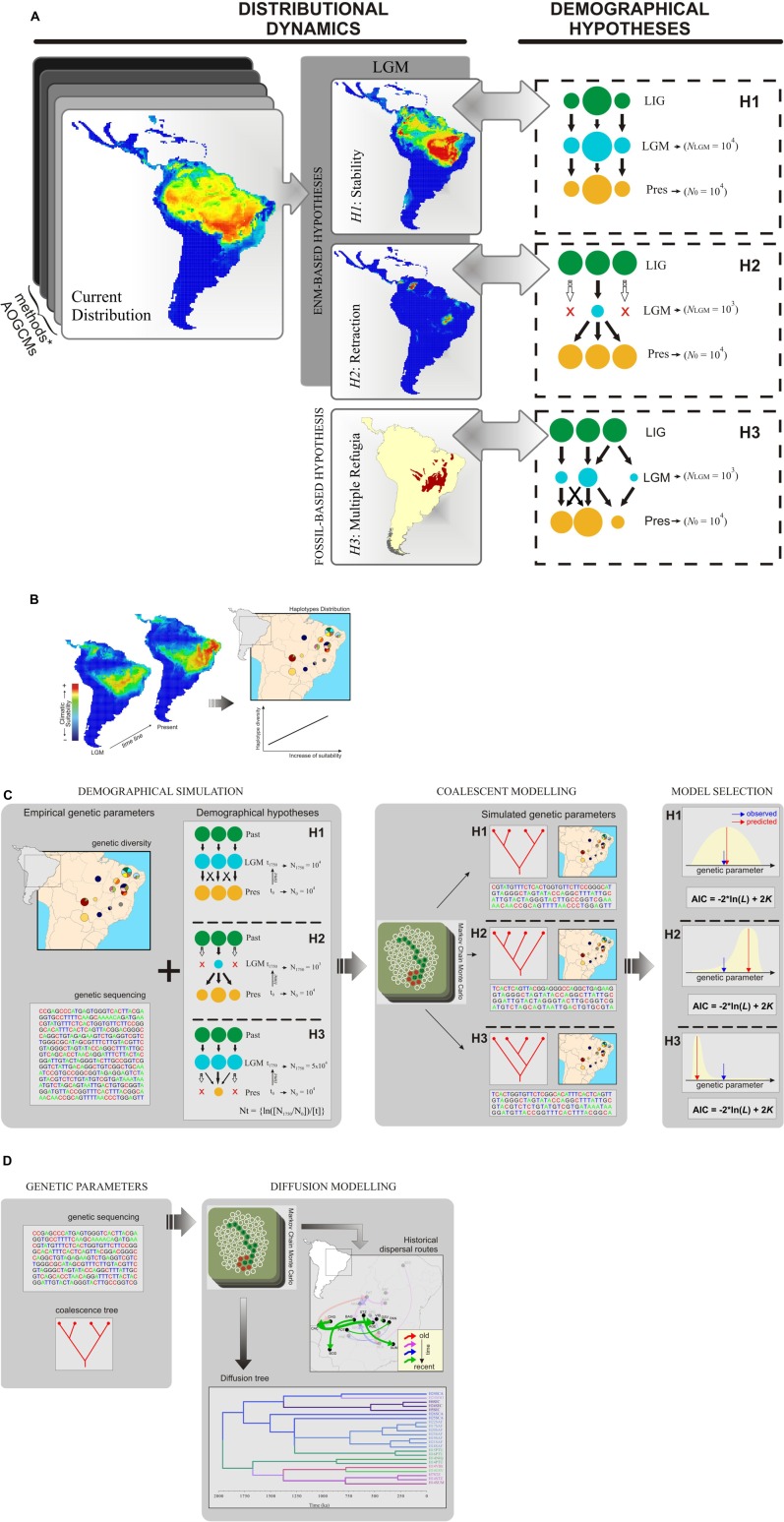
FIGURE 1.
Schematic representation for inferring phylogeographic processes using ecological niche modeling (ENM), coalescence simulation, and relaxed random walk (RRW) model. (A) Uncertainties from ENM methods, climatic simulations (AOGCMs), and other source of evidence (e.g., fossil record) are explored to set multiple distribution dynamics through time, from which alternative demographic hypotheses are inferred. Colored circles represent demes with distinct sizes and locations through time; these demographic scenarios (demographic growth, shrink, and spatial shift) may be simulated using coalescence analyses. LIG: last interglacial (∼125,000 years ago); LGM: last glacial maximum (∼21,000 year ago); Pres: present; NLGM and N0: effective population size at LGM and present, respectively. (B) An ensemble from multiple ENMs provides the environmental context to infer range shifts and perform spatial analyses considering genetic variation across populations (pie charts representing haplotype distributions). Here, maps are representing a geographic range shift of suitable areas toward the northeastern South America from LGM to present-day, which may explain the current spatial pattern of genetic diversity; i.e., increases in climatic suitability across last glacial cycle support higher haplotype diversity at present-day. (C) Coalescent framework used to select the most likely demographic scenario matching the empirical genetic parameters. Demographic hypotheses in a spatially explicit context through time (H1, H2, and H3; colored circles represent the population dynamics trough time) are simulated using coalescent framework to investigate their consequent population genetic structure. Models are selected from multiple criteria, such as likelihood based on posterior predictive distribution and akaike information criterion (AIC). (D) Relaxed random walk (RRW) modeling used to predict historical dispersal routes at the time scale of molecular sequencing data. Saving the time scaling from each model, the dispersal patterns are comparable to the ENM predictions for range shifts (see B). Considering the distribution map from figure, the RRW predicts intermittent dispersal routes through the time in a similar direction of range shift predicted from ENMs. The evidence from multiple models indicates that the dispersal routes predicted by RRW could be the result of climatic forcing across sequential glaciations. Moreover, RRW provides support to explore other features of population dynamics (e.g., source and sink of migrants) and biogeographic processes (e.g., dispersal barriers). Representation of Markov Chain was adapted from Professor Peter Beerli Lecture Notes (http://evolution.gs.washington.edu).