Figure 2.
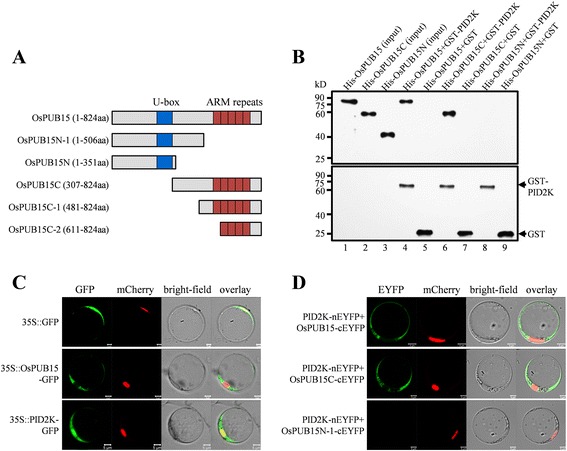
Determination of interactions between OsPUB15 variants and PID2K. (A) Schematic representations of OsPUB15 and its truncated variants. (B) In vitro binding analysis of OsPUB15 and its truncated variants to the PID2 kinase domain. The purified His-fusion proteins were separately mixed with equal quantities of resin bound GST-PID2K and then pulled down by the resin. After extensive washings, the pull-down products were immunoblotted with anti-OsPUB15 (top panel) or anti-GST (bottom panel) antibody. One-tenth of the input of purified His fusions, His-OsPUB15 (lane 1), His-OsPUB15C (lane 2) and His-OsPUB15N (lane 3), were loaded as controls. (C) Subcellular localization of OsPUB15 and PID2K, respectively. Rice protoplasts were co-transformed with the nuclear marker mCherry-VirD2NLS (mCherry-NLS) accompanied with GFP alone (top panel) or GFP fused proteins (middle and bottom panel) driven by 35S promoter, respectively. The panels from left to right are confocal micrographs of GFP signal (green), mCherry signal (red), bright-field images and the resultant overlaid images, respectively. Bars = 5 μm. (D) BiFC analysis of in vivo interaction between PID2K and OsPUB15 or its variants. PID2K and the OsPUB15 variants were respectively fused to the inactive N-terminal (nEYFP) or the C-terminal (cEYFP) part of EYFP, and the pairs of indicated recombinant proteins were transiently expressed in rice protoplasts along with mCherry-VirD2NLS (mCherry-NLS), a nuclear marker. The fluorescence signals were monitored by confocal microscopy. The panel shows fluorescence images of the EYFP signal (green), the mCherry signal (red), the bright-field illumination of protoplasts and the overlaid images, respectively. Bars = 5 μm. The above experiments were repeated three times with similar results.
