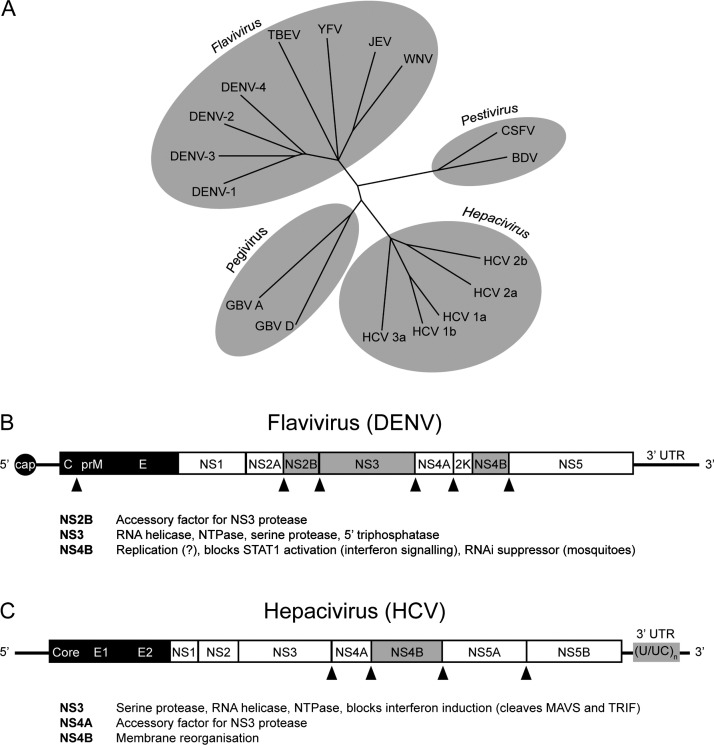
Fig. 2.
Flaviviridae phylogenetic tree and genome organisation. (A) Flaviviridae family tree. Viruses of relevance to this review, and other representative viruses, shown. (B) Mosquito-borne flavivirus (∼11 kb) and (C) hepacivirus (∼9.6 kb) genome organisation (not to scale). Structural proteins are in black. STING antagonists are highlighted in grey. A STING antagonist function has been described for the YFV NS4B protein, but not for the DENV NS4B. Additional known functions for proteins discussed in this review are also given. Triangles indicate cleavage sites for the flavivirus NS2B/3 (B) or hepacivirus NS3/4A (C) proteases. The HCV polyuridine tract ((U/UC)n) is a well-known PAMP that induces type I IFN production. Note that the hepacivirus genome is uncapped. Members of the Flaviviridae lack a poly(A) tail. Viruses not defined elsewhere; CSFV, classical swine fever virus; BDV, bovine diarrhoea virus; GBV, GV virus.