Figure 5.
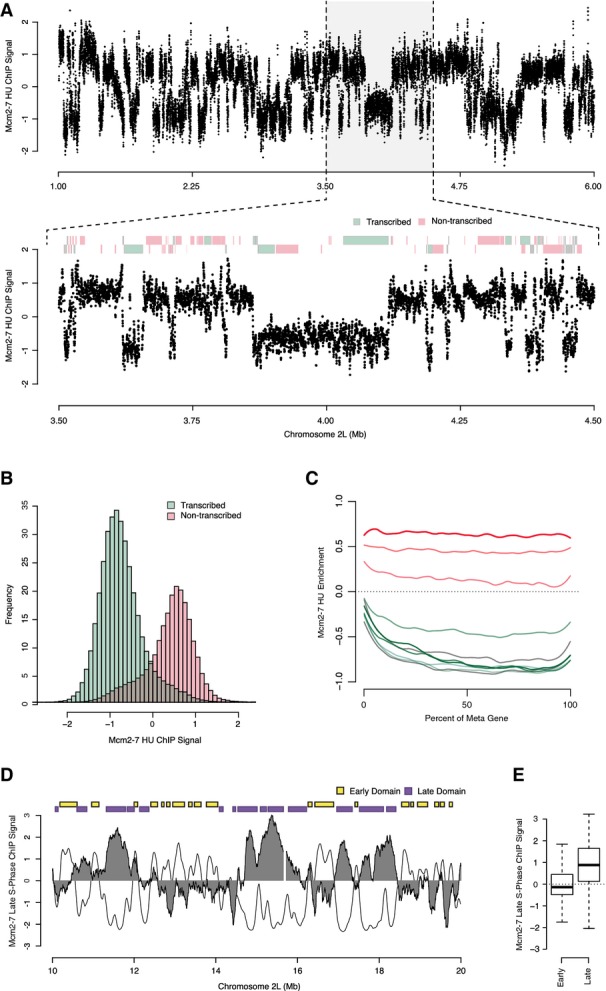
- Genome-wide analysis of Mcm2-7 localization at the G1/S transition by ChIP-chip. Mcm2-7 enrichment from HU-arrested cells is depicted for a 5-Mb section of chromosome 2L. Inset: transcribed (green) and non-transcribed (red) genes are indicated above with genes on the positive strand on the top and those on the negative on the bottom.
- Bimodal distribution of Mcm2-7 enrichment over transcribed and non-transcribed genes. Histogram showing the distribution of probe scores found within transcribed (green) and non-transcribed (red) genes.
- “Meta”-gene analysis of Mcm2-7 enrichment for different deciles of gene expression and their aggregated probe intensities.
- Mcm2-7 is displaced from chromatin by DNA replication. Genome-wide analysis of Mcm2-7 localization in late S-phase (6 h post HU release) by ChIP-chip. Mcm2-7 enrichment is depicted for a 10-Mb section of chromosome 2L (filled gray), replication timing profile (black line) and early (yellow) and late (purple) replication timing domains.
- Box-plots representing late S-phase Mcm2-7 ChIP signal found within early (246) or late (167) replication domains.
