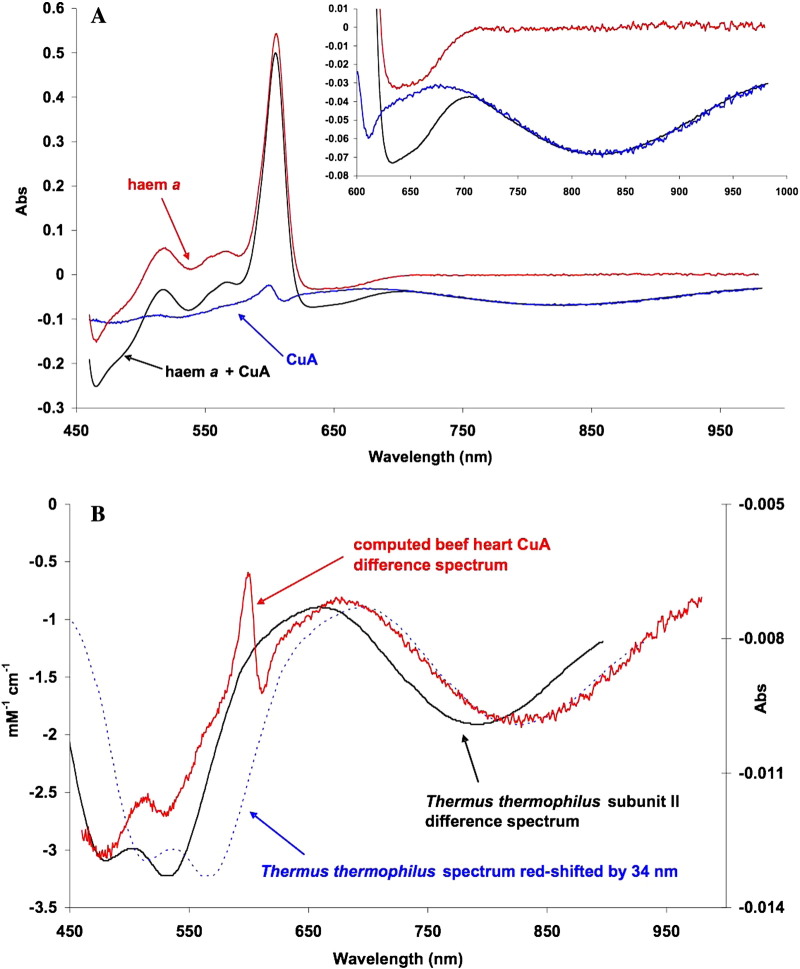
Fig. 3.
Separated redox difference spectra of cytochrome a and CuA.
0.1 M sodium phosphate buffer pH 7.4 0.1% lauryl maltoside 22 °C. with 7.5 mM sodium formate present in the mixed valence sample (haem a CuA reduced/haem a3 CuB oxidised). Formate complexes formed as described in Materials and methods.
A. Separated difference spectra +/− formate (haem a and CuA):
------ formate mixed valence minus oxidised formate (haem a Fe2 + plus CuA+) (IV− VIII, Table 2);
 formate mixed valence minus calculated CuA (haem a Fe2 +); (i.e. haem a reduced minus oxidised).
formate mixed valence minus calculated CuA (haem a Fe2 +); (i.e. haem a reduced minus oxidised).
 formate mixed valence minus calculated haem a (CuA+) (i.e. CuA reduced minus oxidised).
formate mixed valence minus calculated haem a (CuA+) (i.e. CuA reduced minus oxidised).
B. Bacterial and mammalian CuA reduced minus oxidised difference spectra.
 CuA difference spectrum obtained by reduction in the presence of formate (converted to extinction coefficients);
CuA difference spectrum obtained by reduction in the presence of formate (converted to extinction coefficients);
-------- Thermus thermophilus subunit II difference spectrum (digitised version from the cytochrome oxidase Web site http://www-bioc.rice.edu/~graham/CcO.Spectra.pc).
The troughs in the visible spectra are closely matched but in the NIR the bacterial subunit II spectrum is blue shifted by ~ 34 nm relative to the computed cytochrome c oxidase CuA . The 599 to 611 nm spike is due to the haem a spectral shift that occurs upon CuA reduction (see Discussion).