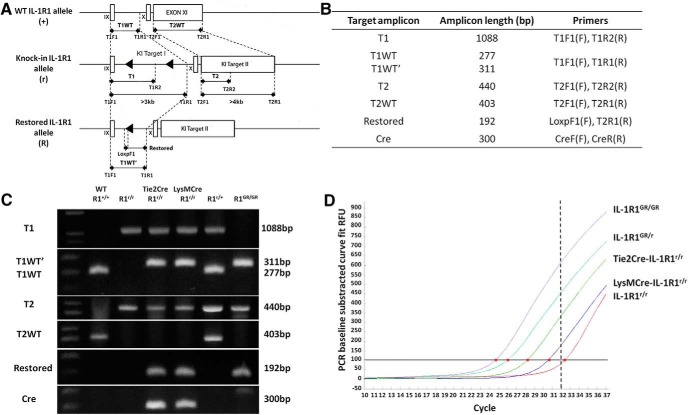
Figure 3.
Primer design for detection of knock-in allele and genotyping identification. A, Top, Genomic organization of exon IX, X, and XI in the wild-type IL-1R1 allele. Middle, Knock-in IL-1R1 allele after homologous recombination. Bottom, Restored IL-1R1 allele after Cre-recombinase-mediated excision of the disrupting sequence. Black triangles indicate loxP sequence and primer design is shown by black arrows. Sequence length represented by bars in the schematic has been adjusted for better illustration. Dashed lines indicate primer locations in the allele. B, Table of designations of target amplicons, their corresponding sizes, and primers. C, Genotyping results of animals carrying different IL-1R1 alleles. D, SYBER green quantitative RT-PCR with primers LoxPF1 and T1R1 on 1 μg of mouse tail DNA from animals with different genotypes. Different amount of the Restore amplicon were detected in the same amount of total genomic DNA from either an IL-1R1GR/GR animal or a cell-type-specific IL-1R1 restored animals. Red dots indicate the coordinate (amplification cycle number) of the amplification curve that crossed the threshold of SYBER green detection level. The threshold detection level of specific amplification of our real-time PCR machine is 32 cycles as shown by the dashed line.