Figure 3.
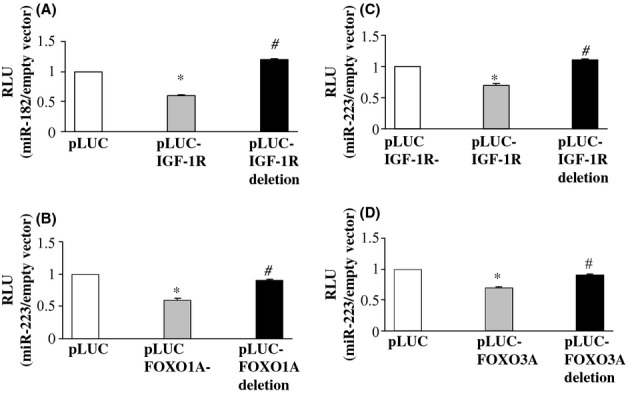
Functional binding assays. (A) Binding of miR-182 to the 3′UTR region of IGF-1R. (B) Binding of miR-223 to the 3′UTR region of FOXO1A. (C) Binding of miR-223 to the 3′UTR region of IGF-1R. (D) Binding of miR-223 to the 3′UTR region of FOXO3A. Each pLUC vector contains the CMV promoter, the firefly luciferase gene, the 3′ UTR of a target gene, and a SV40 terminator sequence. When a miRNA is expressed and binds to the 3′ UTR, it results in repression of luciferase gene expression. We used pLUC plasmids encoding for the 3′-UTR regions of the mRNA targets IGF-1R, FOXO1A, and FOXO3A (pLUC IGF-1R, pLUC FOXO1A, and pLUC FOXO3A) and for the 3′-UTR regions of the same mRNA targets containing deletions (pLUC IGF-1R deletion, pLUC FOXO1A deletion, and pLUC FOXO3A deletion). Each pLUC vector was co-transfected into the HEK293T cells with a plasmid-encoding Renilla luciferase along with a plasmid-encoding miR-182 or miR-223 or the empty vector. Also pSUPER-scramble oligos were used for co-transfection into the HEK293T cells. No significant differences were observed when pSUPER-scramble oligos were used instead of empty vector. Firefly luciferase values were normalized to Renilla luciferase activity, and the ratio of luciferase activity of each construct in the presence or in the absence of exogenous miR-182/-223 was calculated. Data are reported as relative light units (RLU) and in relation to empty vector. Mean value ± SD of three independent experiments are shown. *P ≤ 0.05 pLUC-target vs. pLUC; #P ≤ 0.05 pLUC-target-deletion vs. pLUC. (For details see Experimental procedures).
