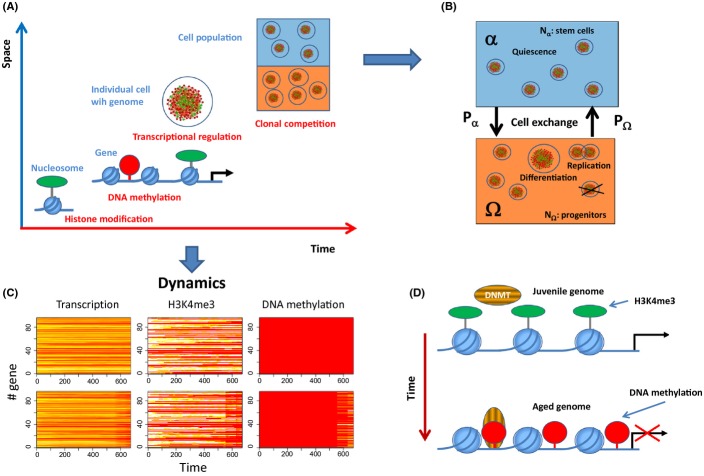
Figure 1.
Multi-scale model of epigenetic drifts in aging cells. (A) Time and length scales covered by our model. The basic unit of the model is the nucleosome, the histones of which undergo H3K4 (de-) modification. The average H3K4me3 level controls the access of CpG-methylating enzymes to nearby CpG sites. Both H3K4 and DNA methylation interfere with the transcription factor-network of the cell that is defined by its genome. All cells are equipped with the same genome but underlie individual aging and thus reside in individual regulatory states. Aging depends on signals provided by the environment. We consider two environments α and Ω. (B) In the α environment mimicking a stem cell niche (blue) cells are quiescent, while in the Ω environment (orange), they can proliferate and differentiate with rate R and q, respectively. Differentiated cells are removed from the system. Cells make transitions between α and Ω with probabilities Pα and PΩ. (C) Regulatory profiles of individual cells: Transcription, H3K4me3 modification, and DNA-methylation profiles (red-yellow-white: low–high values) for a cell fixed in environment α (upper row) and Ω (lower row). H3K4me3 modification and DNA methylation are given as the fraction of modified nucleosomes and methylated CpG sites associated with the gene, respectively. Time is given in computational time steps Δt. Proliferation is required for changes in DNA methylation, while changes of H3K4me3 modification occur also in quiescent cells. (D) Age-dependent phenotypic changes. Initially the nucleosomes carry H3K4me3 modifications which protect the associated DNA from becoming methylated. Over time this modification is lost enabling methylation of the nearby CpG sites and stable silencing of the associated gene.