Figure 1.
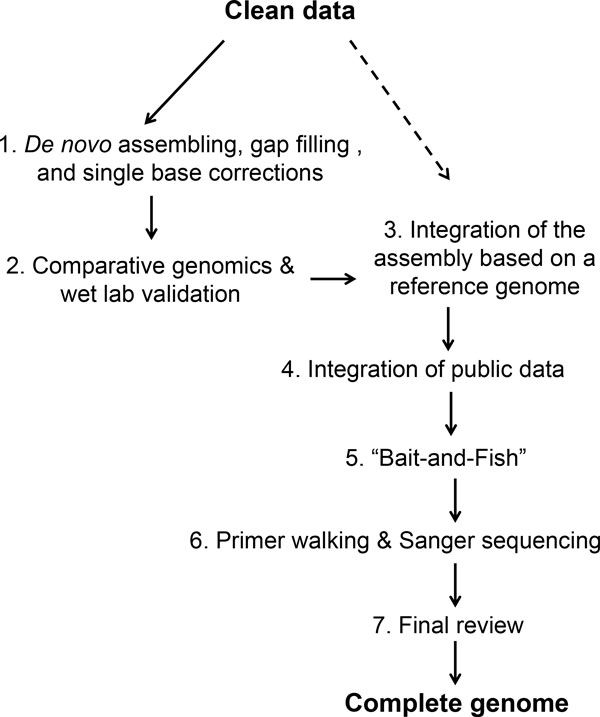
The pipeline of genome assembling and gap closure. Clean data were subject to de novo assembling, initial gap filling, and single base corrections with SOAPdenovo and SOAPaligner; the resulting assembly was used for comparative genomics studies and also provided guidance for wet lab validations. Meanwhile, the clean data were assembled separately based on a reference genome using CLC Genomics Workbench. This second assembly was integrated into the first one to yield a hybrid assembly, which was then updated with public data, GapFish results, and Sanger sequencing data until the genome sequence was complete.
