Figure 4.
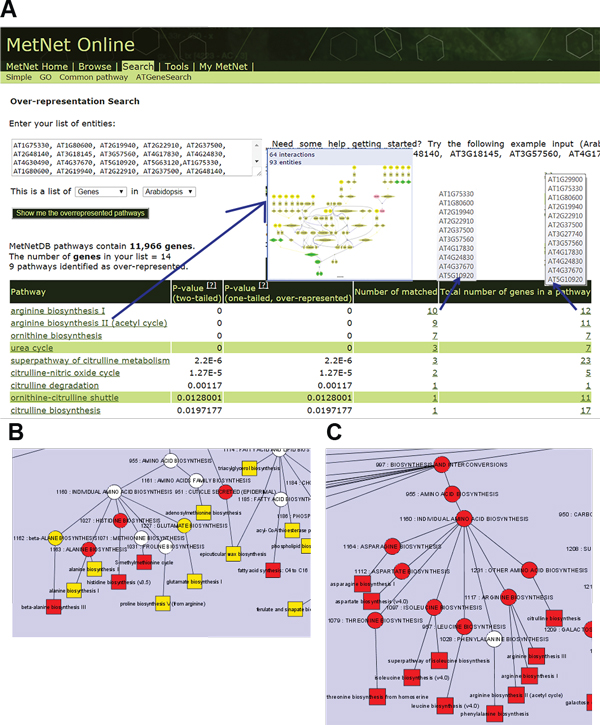
Co-analysis of transcriptomic and metabolomic data of developing seeds. A. Fisher exact test analysis of the differentially accumulated transcripts (p-value< 0.05) using MetNet showing pathways that are over-represented. Clicking on or mousing-over a pathway brings up the pathway map, and the lists of locus IDs involved in that pathway. B. MRPP test [61] analysis of transcriptomics data showing amino acid biosynthesis pathways that are differentially expressed across the developmental time course. C. MRPP test analysis of metabolomics data showing amino acid biosynthesis pathways that exhibiting significant changes in accumulation patters over the developmental time course. Oval: category of pathways; square: single pathway. Colors: q-value < 0.01, red; q-value < 0.05, orange.
