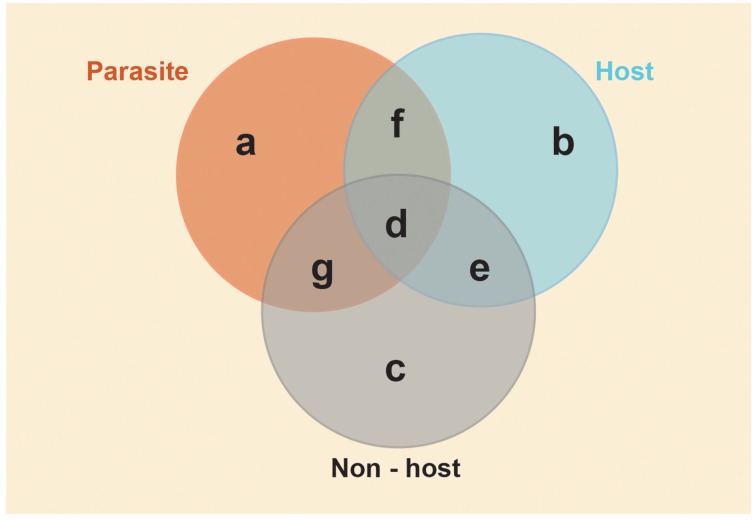
Figure 2.
Conceptual framework for predictions on shared and contrasting genomic/phenotypic diversity in social parasite/host relationships. Venn diagram depicting predicted shared and contrasting molecular phenotypes of non-hosts, hosts and social parasites. We define the molecular phenotype to include contrasting patterns of gene expression (significant up or down regulation), gene regulatory elements (e.g., non-coding RNAs, microRNAs, DNA methylation, histone modifications), gene interaction networks (e.g., correlated co-expression) and protein synthesis. Each area represents the molecular phenotype of the specific suite of traits. Overlapping areas indicate putatively shared molecular phenotypes. The yellow shaded area indicates the shared environment of the three species, which we predict will cause similar responses in molecular phenotypes of all three species. Conserved generic traits (area d): Molecular processes underlying traits shared by all species, and thus putatively inherited from their common ancestor. These will include fundamental machinery for growth, cell repair, metabolism, as well as more specific traits of interest that are shared among queens and social parasites such as aggression and reproduction. Identifying the molecular phenotype of this area allows tests of the genetic toolkit hypothesis. Parasite-specific (area a): Molecular processes underlying traits that have evolved in the parasite to facilitate its specialized parasitic life style, for example enhanced fighting ability, usurpation behaviors, cryptic mimicry. Identifying the molecular phenotype of this area addresses the question of whether newly evolved phenotypic traits require new genes/pathways or simply re-use existing ancestral genes/pathways. Free-living specific (area e): Molecular processes underlying free-living traits that no longer provide a fitness advantage to social parasites, e.g., parental care traits and nest founding. Identifying the molecular phenotype of this area allows us to determine what happens at the molecular level when phenotypic traits are lost, e.g., are there changes in regulation/expression, loss of processes/genes? Restricting this to traits/genes shared by free-living host and non-host species is likely to represent the traits present in the eusocial ancestor of the social parasite, and exclude processes that may have evolved subsequently. These latter processes may be associated with social parasite resistance (areas c and g) in sympatric non-hosts, host response to parasitism (area b) and co-evolved traits (area f) in host and parasite that are absent from the non-host.