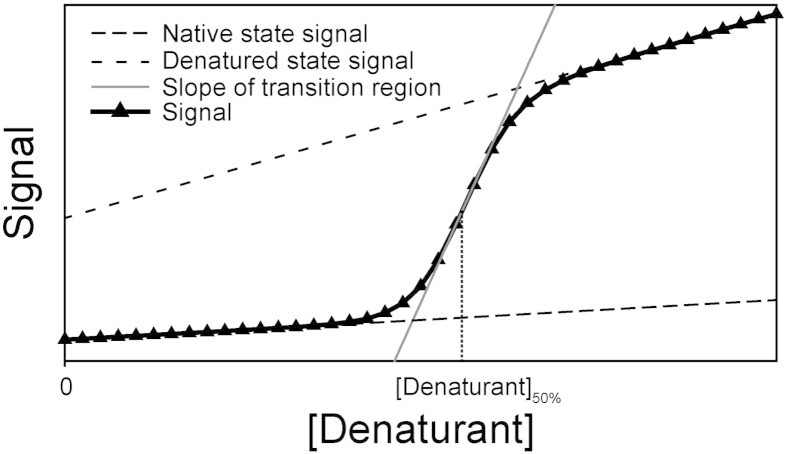
Fig. 1.
Chemical denaturation of a protein at equilibrium. Simulated 2-state denaturation induced by increasing the chemical denaturant concentration. The spectroscopic signal used to probe denaturation is typically sigmoidal (black triangles) and consistent with cooperative unfolding [13,14,16]. In order to determine the fractional occupancy of native and denatured states, it is necessary to fit the slopes of pre- (native) and post-transition (denatured) baselines to linear functions (dashed lines). The ΔGD−N0 value can be easily determined by linear extrapolation once the transition midpoint ([Denaturant]50%) and mD–N values are known (see Eq. (1), see Section 1.1). mD–N correlates with the slope of the transition (grey line) and increases with protein size (Fig. S1b) [13,23].