Abstract
Background
46,XY sex reversal is a rare disorder and familial cases are even more rare. The purpose of the present study was to determine the molecular basis for a family with three affected siblings who had 46,XY sex reversal.
Methods
DNA was extracted from three females with 46,XY sex reversal, two normal sisters, and both unaffected parents. All protein coding exons of the SRY and NR5A1 genes were subjected to PCR-based DNA sequencing. In addition, array comparative genomic hybridization was performed on DNA from all seven family members. A deletion was confirmed using quantitative polymerase chain reaction. Expression of SOX9 gene was quantified using reverse transcriptase polymerase chain reaction.
Results
A 349kb heterozygous deletion located 353kb upstream of the SOX9 gene on the long arm of chromosome 17 was discovered in the father and three affected siblings, but not in the mother. The expression of SOX9 was significantly decreased in the affected siblings. Two of three affected sisters had gonadoblastomas.
Conclusion
This is the first report of 46,XY sex reversal in three siblings who have a paternally inherited deletion upstream of SOX9 associated with reduced SOX9 mRNA expression.
Keywords: XY sex reversal, gonadal dysgenesis, SOX9, SRY, NR5A1
1. INTRODUCTION
Disorders of sex development are uncommon. 1 in 300 newborn babies have some developmental abnormality to cause concern and prompt further investigation. It is estimated that 1 in 4500 newborns have a serious abnormality of the external genitalia whereby the sex of the baby cannot be determined by examination alone. (1). Even more rarely, children are born with normal male or female genitalia and are raised appropriate for the sex of the external genitalia, but are determined to have complete sex reversal (either 46,XY female or 46,XX male) at a later stage in their life.
Mutation in the SRY gene was first reported as a cause for 46,XY sex reversal in 1990(2). Subsequently multiple case reports of the involvement of SRY in 46,XY sex reversal have been published (3). It is estimated that about 10–15% of cases of 46,XY sex reversal can be explained by mutations in SRY gene(1, 4). Mutations in NR5A1 were first reported to cause 46,XY sex reversal in 1999(5). Single gene mutations other than in the above two genes are considered rare causes for 46,XY sex reversal (4).
Most cases of 46,XY sex reversal are sporadic and isolated. Although rare, cases of familial 46,XY sex reversal have been reported in the literature (6). It has been recognized that these rare cases of familial 46,XY sex reversal may be useful in furthering our understanding of the causes of sexual development. Decreased DNA binding activity of SRY protein with variable penetrance, mosaicism in peripheral blood, and germ cell mosaicism are some of the factors that have been shown to be involved in the occurrence of familial 46,XY sex reversal (6).
The SRY related SOX9 gene is involved in sex determination. Mutations in this gene typically cause campomelic dysplasia in addition to sex reversal(7). A small proportion of patients develop acampomelic dysplasia(8). Here we report a family of five siblings who are all phenotypically female. Three of these females are karyotypically 46,XY and SRY positive. The purpose of the present study was to identify the molecular basis of 46,XY sex reversal in this family.
2. MATERIALS AND METHODS
2.1 PATIENTS
The index case III3 was a 19 year old Mexican female who presented with a left adnexal mass and primary amenorrhea (Figure 1). She was 5 feet 0 inches tall (and attained a final height of 5 feet 7 inches), and had Tanner stage 2 breasts with Tanner stage 2 pubic hair. Normal female external genitalia without clitoromegaly, a small cervix, and a large pelvic mass were observed on physical exam. Serum FSH and LH were elevated at 49.5 mIU/mL and 27.4 mIU/mL initially and on several repeat occasions. CT scan of the abdomen and pelvis revealed a large (11×7.4×6.6 cm) mass in the cul-de-sac, an atrophic uterus, and normal kidneys bilaterally. A pre-operative pericentromeric FISH, prompted by the amenorrhea, revealed one X and one Y in each of the 200 cells examined, and a full karyotype revealed 46,XY in 20 cells.
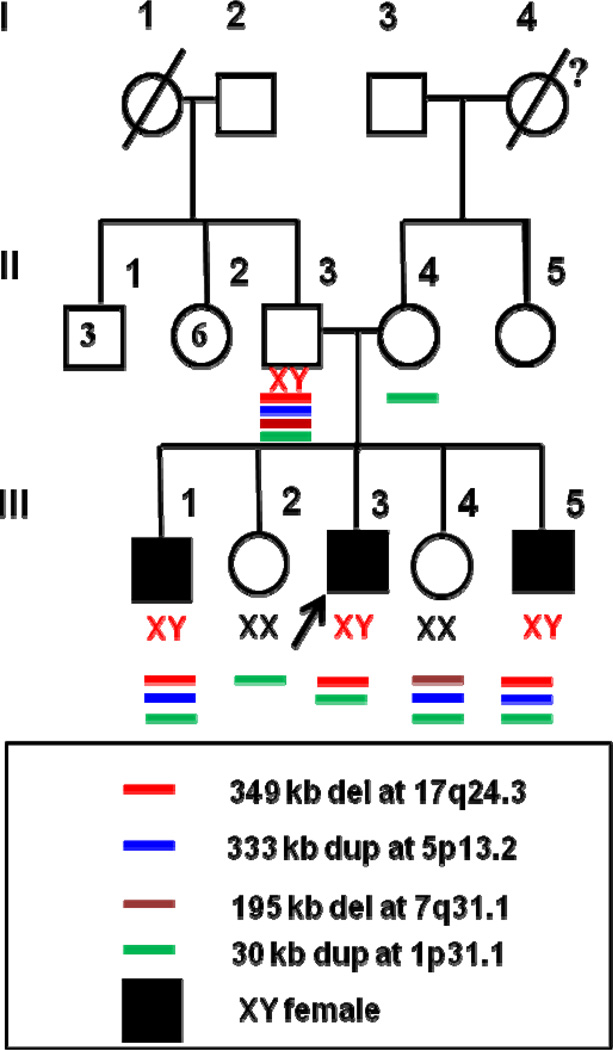
Figure 1.
Family pedigree and the associated deletions and duplications identified by array CGH. 46,XY sex reversal patients are indicated by shaded squares. The arrow indicates the index case. Roman numerals indicate the different generations. The numbers within the square and circle in generation II indicate the number of brothers and sisters of the index case’s father.
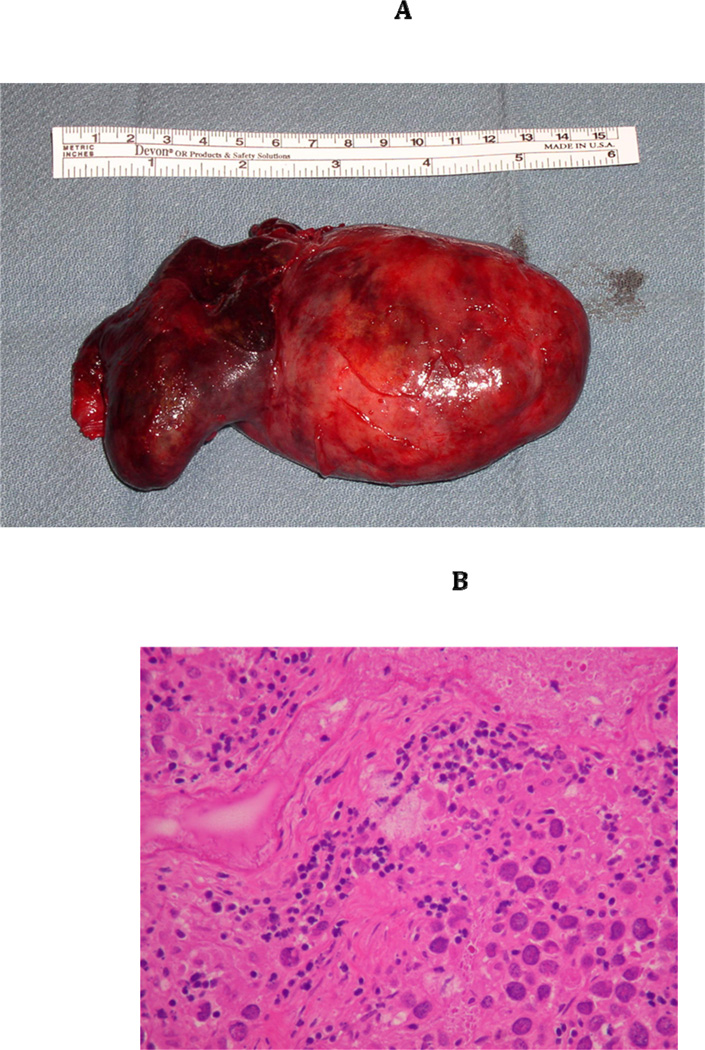
By laparotomy, a 10 cm left adnexal mass was removed, which was reported to be a dysgerminoma. Lymph nodes were negative for metastasis. The right streak gonad, which was also removed , was noted to have a small focus of gonadoblastoma as well as dysgerminoma (Figure 2A–D). The uterus appeared small but normal. She underwent three cycles of chemotherapy for stage IB ‘ovarian’ cancer. III3 was otherwise healthy and had no medical problems. She had an X-ray of the femur that showed no significant bowing.
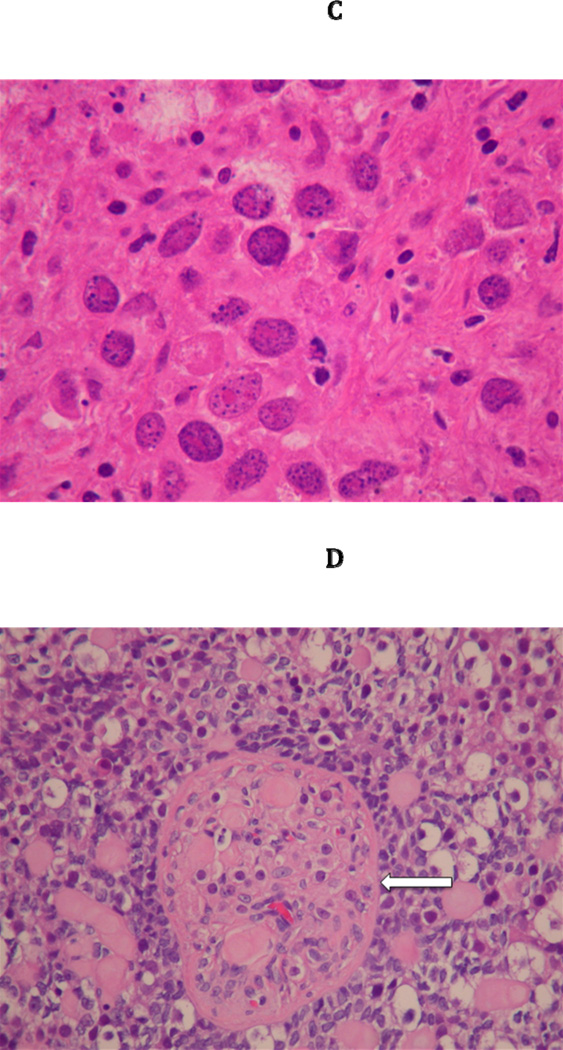
Figure 2.
(A) Gross Pathology of left gonad of III3 with 46,XY sex reversal. (B) Medium power view of the left dysgerminoma from III3 with the lymphocytic infiltrate characteristic of dysgerminoma/seminoma. (C) High power view, representative image of the 9 × 7 × 4 cm, almost completely necrotic left ovarian mass from III3. The cells are undergoing coagulative necrosis. Note the nucleolar prominence. (D) Right gonad from III3 with 0.4 cm focus of gonadoblastoma (White Arrow). Note nests of tumor cells composed of large germ cells with clear cytoplasm and smaller sex-cord type cells that surround the germ cells. Deposits of eosinophilic basement membrane material can be seen. Within this gonad, a 3 mm area of dygerminoma was also found (not shown).
Patient III3 had 4 siblings who were all female and were karyotyped—III1 (23 yrs), III2 (22yrs), III4 (17 yrs) and III5 (15 yrs). Older sister III1 (23 years) was a healthy, phenotypic female of 5 feet 6 inches height who had Tanner 1 breasts, primary amenorrhea, and a 46,XY karyotype (20 cells). No bone deformities were noted. A pelvic sonogram revealed a small uterus (5.3×1.6×2cm) with a 1.3×1.1cm right gonad; the left gonad was not visualized. She underwent laparoscopic bilateral gonadectomy. Histopathological examination revealed bilateral streak gonads. III2 (22 years, 5 feet 2 inches tall) and III4 (17 years, 5 feet 3 inches tall) were healthy females who had attained menarche at 12 to 13 years age, were without bone deformities, and were both 46,XX. III2 subsequently gave birth to a normal male child and III4 to a normal son and a normal appearing daughter. III5 (15 years) had Tanner stage 1 breasts, primary amenorrhea, and a 46,XY karyotype [20 cells]. She was 5 feet 9 inches tall and had no bone deformities. A pelvic sonogram revealed a small uterus (3.9×1.2×1.7cm) and neither gonad was visualized. She underwent a laparoscopic bilateral gonadectomy. Histopathological examination revealed a streak gonad on the right and a small gonadoblastoma on the left.
In this Mexican family (Figure 1), both parents attained puberty at a normal age and were not related. There is no history of bone deformities in either parent. Her mother had one sister who had normal children. The proband’s maternal grandfather was alive at 72 years age, but the maternal grandmother died of unknown cause. The proband’s father had six sisters and three brothers. The paternal grandfather was alive at 92 years age and the grandmother was deceased secondary to complications from diabetes. There were no reported abnormalities in the extended family, including bone deformities. All 46,XX females apparently attained menarche at a normal age.
2.2. METHODS
The Institutional Review Board approvals were obtained and each patient signed a consent. Blood was drawn from all seven subjects for genomic DNA extraction from the white blood cells using previously described techniques(9). The single exon SRY gene (NM_003140.1) was amplified as one fragment by PCR and sequenced using the BIG-DYE Termination method on an ABI Prism 310 automated DNA sequencer (Applied Biosystems, Foster city, CA). Similarly, the protein coding exons (exons 2–7) and the flanking splice sites of NR5A1 (NM_004959.4) were subjected to PCR-based DNA sequencing(10). DNA sequence from this family was compared to the SNP database and 1,000 Genomes Database to exclude polymorphisms.
All seven subjects were also tested for copy number variants (CNV) by array comparative genomic hybridization with the use of Agilent 244K arrays (Santa Clara, CA) as described previously(11), and compared with the Datsbase of Genomic Variants to exclude known copy number variants known to occur in healthy population. These arrays contain 244,000 CNV probes spaced with three unique probes per 20kb distance. Quantitative PCR was performed on genomic DNAs from all seven subjects and one control male to confirm the deletion upstream of SOX9. The experiments were performed three times to confirm reproducibility and to account for variation. Primers used for the amplification of the deleted region are following: forward 5’ – TCTAGGGT ACACTCCTGTTAGG-3’ and reverse 5’ – GAAGGTGAGGGTCTCTGTAATC-3’. A primer set for the amplification of GAPDH exon 5 region was used for the internal control: forward 5’ – GATCATCAGCAATGCCTCCT – 3’ and reverse 5’ – ATGGCATGGACTGTGGTCAT – 3’. A total of 10 ng of genomic DNA was mixed with Power SYBR Green PCR Master Mix (Applied Biosystems, Carlsbad, CA) and primers. Quanitative PCR was performed for the amplification of the targeted region utilizing a 7500 Real-Time PCR system (Applied Biosystems, Carlsbad, CA). The PCR reactions were cycled 40 times after initial denaturation (95°C, 10 min) with the following parameters: denaturation at 95°C for 15 seconds and annealing/extension at 60°C for 1minute. The levels of GAPDH amplification were used to normalize relative levels of DNA.
Transcription of SOX9 was then quantified on lymphoblast RNA reverse transcription-quantitative PCR (RT-qPCR) from all seven subjects and one control male. The experiments were performed three times to confirm reproducibility and to account for variation. Total RNA was extracted from the pellet using RNeasy Mini Kit (Qiagen, Valencia, CA) according to the manufacturer's protocol. RNA (2 µg) was reverse transcribed and converted into complementary DNA (cDNA) using the RevertAid First Strand cDNA synthesis kit (Thermo Scientific, Waltham, MA). The following are primer sequences used for amplification of SOX9 and GAPDH; SOX9 forward 5’ – TCTGGAGACTTCTGAACGAGAG – 3’ and reverse 5’ –CGCGGCTGGTACTTGTAATC -3’; GAPDH forward 5’ – GATCATCAGCAATGCCTCCT – 3’ and reverse 5’ –ATGGCATGGACTGTGGTCAT – 3’. The cDNAs were mixed with Power SYBR Green PCR Master Mix (Applied Biosystems, Carlsbad, CA) and primers. All samples and loading controls were plated in triplicate and centrifuged briefly. RT-qPCR was performed for the amplification of SOX9 and GAPDH utilizing a 7500 Real-Time PCR system (Applied Biosystems, Carlsbad, CA). The PCR reactions were cycled 40 times after initial denaturation (95°C, 10 min) with the following parameters: denaturation at 95°C for 15s and annealing/extension at 60°C for 1min. Expression of GAPDH was used to normalize relative expression of SOX9 mRNA. ΔΔCT was automatically calculated by the 7500 Real-Time PCR system(12).
The method of linear contrasts was used to compare the mean expression of SOX9 in the control with that in the father, the three XY siblings (III1, III3, III5) treated as a group, the two unaffected siblings (III2, III4) treated as a group, and the mother(13, 14). Adjustment was made for the fact that there were replicated observations for each subject. All analyses were performed using SAS 9.3 (SAS Institute Inc., Cary, NC, 2010).
3. RESULTS
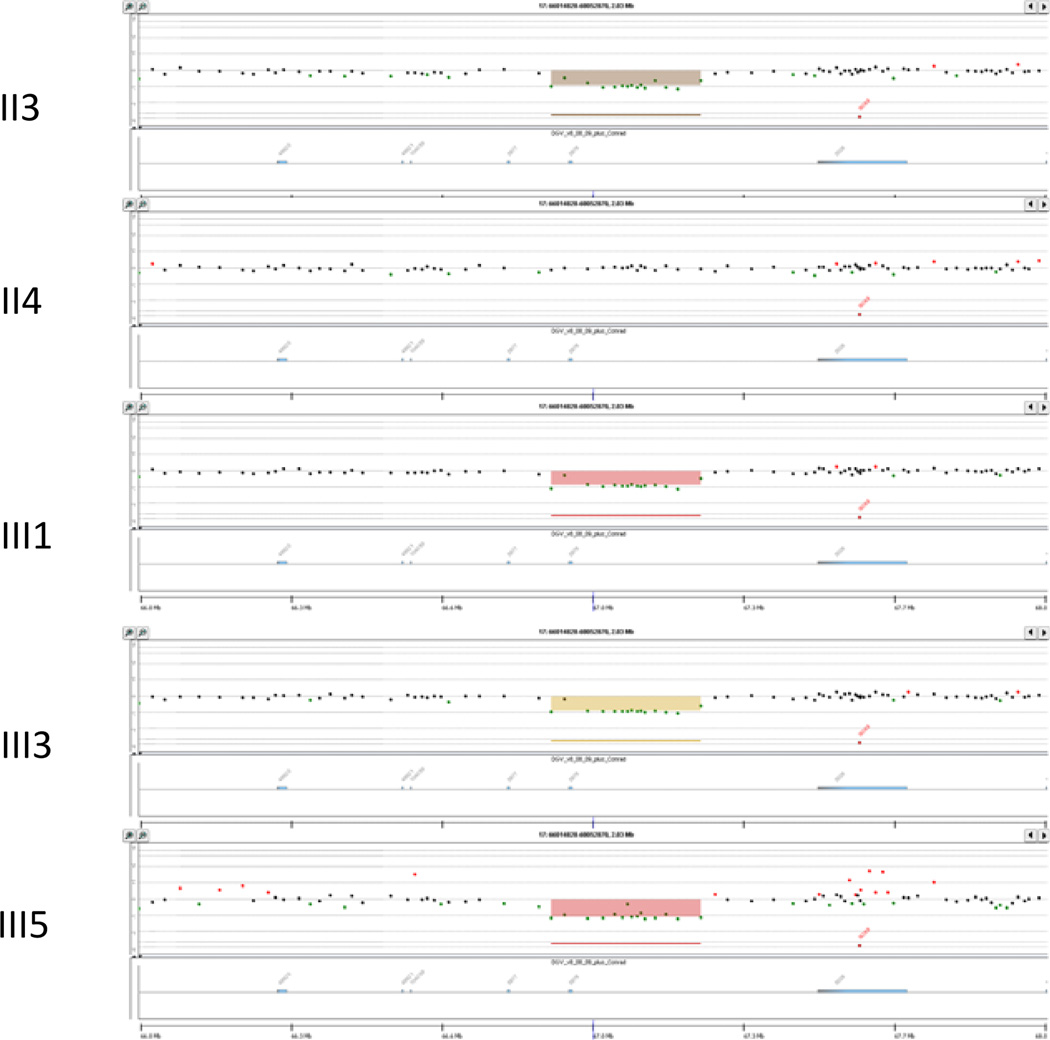
No exonic deletions were identified in any of the subjects or unaffected family members. SRY sequence was normal in the father and the three 46,XY sex reversed siblings. NR5A1 sequence was normal in all three affected 46,XY sex reversal patients., Array CGH revealed a 349 kb deletion at 17q24.3, which is 353 kb upstream of SOX9. This deletion was observed in the father and all three XY siblings (III1, III3 and III5), but not in the two XX siblings (III2 and III4) or mother (Figure 3). In addition, III1 (XY), III4 (XX) and III5 (XY) had a 333 kb duplication at5p13.2, which was inherited from their father. III4 (XX) had a 195 kb deletion at 7q31.1 inherited from her father. All five siblings had a 30 kb duplication at 1p31.1 seen in both the father and the mother. The mother had no other deletions or duplications detected. Except for the 349 kb deletion, none of the other CNVs segregated with 46,XY sex reversal in this family (Figure 1).
Figure 3.
A 349 kb deletion at 17q24.3 involving the father (II3) and three affected siblings (III1, III3, III5), but not the mother (II4) or the two 46,XX siblings (not shown).
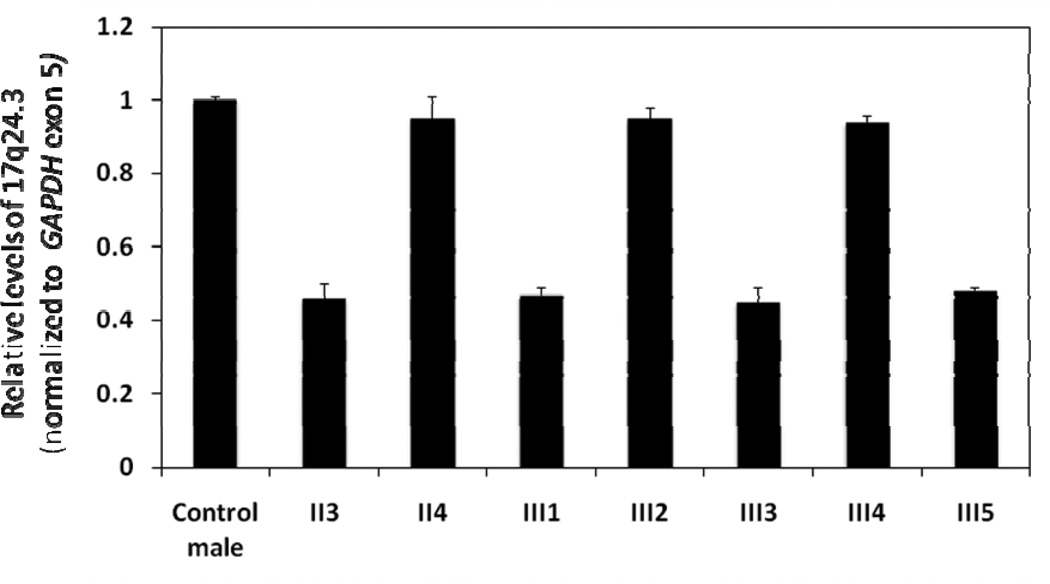
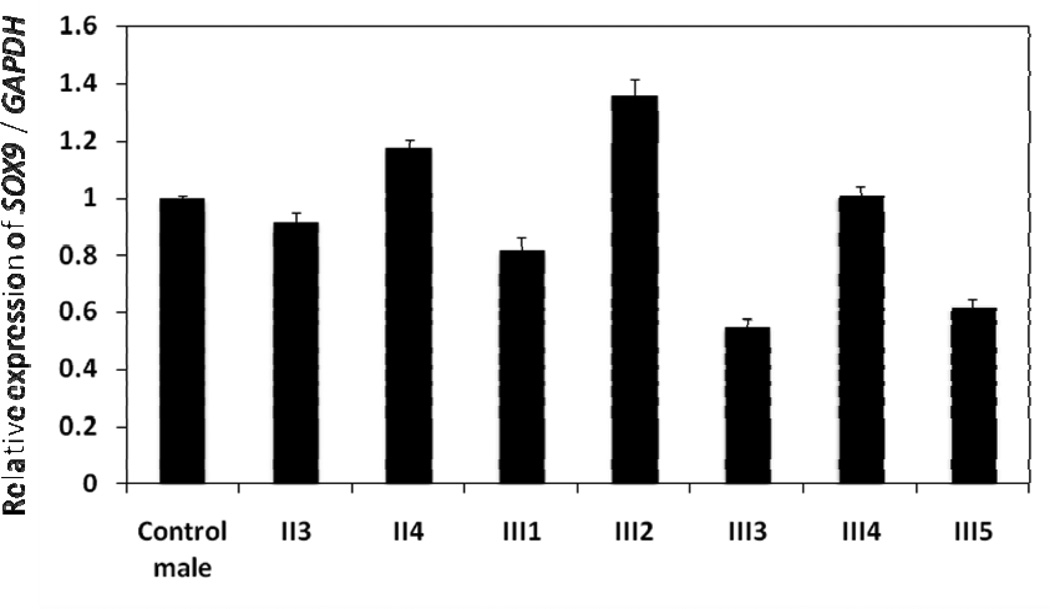
Quantitative PCR of genomic DNA confirmed the deletion upstream of SOX9 in the father and all three XY siblings (III1, III3 and III5) (Figure 4). The PCR products were approximately 50% reduced compared with the control male, the mother, and the two unaffected siblings (III2 and III4). RT-qPCR demonstrated reduced mRNA expression of SOX9 in all three XY siblings (III1, III3 and III5) compared to the control and the father (Figure 5). Transcript levels were also decreased compared with the mother and the two unaffected siblings (III2 and III4). Table 1 contains summary statistics for the mean expression of SOX9 in each of the eight study subjects. Table 2 contains a summary of the linear contrast analyses comparing the control with the father (II3), the three XY siblings treated as a group, the two unaffected siblings treated as a group, and the mother. As indicated, the father did not differ significantly from the control. The overall mean expression for the three XY siblings was significantly lower than the control (p < 0.0001), whereas the overall mean expression for the two unaffected siblings was significantly higher than the control (p = 0.0002). Mean expression of SOX9 in the mother was also significantly higher than the control (p = 0.0019).
Figure 4.
Quantitative PCR performed on genomic DNA to confirm del17q24.3 in the father (II3) and the three affected siblings (III1, III3, III5) compared with a control male and 46,XX members of the family. See Materials and Methods for details.
Figure 5.
RT-qPCR of SOX9 expression in the family and a control male. See Materials and Methods for details.
Table 1.
Summary Statistics for Mean Expression of SOX9 in Eight Study Subjects
| Subject | n | Mean | Standard Deviation |
Minimum | Maximum |
|---|---|---|---|---|---|
| Control | 9 | 1.001 | 0.040 | 0.95 | 1.07 |
| Father (II3) | 9 | 0.949 | 0.106 | 0.81 | 1.15 |
| Mother (II4) | 9 | 1.182 | 0.088 | 0.98 | 1.29 |
| III1 | 9 | 0.819 | 0.139 | 0.66 | 1.00 |
| III2 | 9 | 1.369 | 0.185 | 1.13 | 1.62 |
| III3 | 9 | 0.612 | 0.127 | 0.45 | 0.80 |
| III4 | 9 | 1.013 | 0.086 | 0.87 | 1.15 |
| III5 | 9 | 0.673 | 0.122 | 0.56 | 0.92 |
Table 2.
Summary of Results for Linear Contrasts of Mean Expression of SOX9 vs. Control
| Individual or Group |
Mean Difference |
Standard Error | t | d.f. | p-value |
|---|---|---|---|---|---|
| Father (II3) | −0.052 | 0.045 | −1.16 | 16 | 0.2638 |
| Mother (II4) | 0.181 | 0.045 | 4.02 | 16 | 0.0010 |
| XY siblings (III1, III3, III5) | −0.300 | 0.037 | −8.14 | 16 | < 0.0001 |
| Unaffected Siblings (III2, III4) | 0.190 | 0.039 | 4.87 | 16 | 0.0002 |
4. DISCUSSION
The genetic cause for sex reversal can be identified in approximately 50% of cases. Mutations in SRY are present in about 10–15% of cases (4), and there are case reports of SRY mutations being inherited from the father(6, 15). Therefore, although the father was not affected, SRY was first analyzed and determined to be normal in all the family members with a Y chromosome. Initial reports of an association between mutations in NR5A1 and sex reversal were described in patients with coexistent adrenal dysfunction (5). A mutation in NR5A1 was later identified in approximately 15% of sex reversal patients without any adrenal involvement (10). Therefore, NR5A1 was sequenced, but somewhat surprisingly (given the phenotypic normalcy of the affected siblings), no mutations were identified.
Additional candidate genes such as WT1 (16) or SOX9 (7, 17) also play a major role in the sex determination pathway., It was also apparent that many other genes (4) including GATA4 (18), DHH (19, 20), CBX2 (21) and ATRX (22) should be considered and therefore, at this point in the investigation, an array CGH was used to screen the genome for CNVs. Having excluded a mutation in SRY, it was hypothesized (just as Benko et al (23) did) that the mutation must be maternally inherited with a male ‘permissive’ gene to have affected the children but spared the father. It was a surprise therefore to find the deletions in the father and affected siblings. The deletion on 17q24.3, which is 353 kb upstream of SOX9, could explain the phenotype as it was present in the father, all three affected sex reversal patients, and in none of the unaffected members (mother and normal XX sibs).
In the family described in our study, several CNVs were identified. Interestingly they were all inherited from the father to his children except the 1p31.1 duplication of 30 kb, which was found in both the mother and father. Of the candidate regions, the 333kb duplication in chromosome 5p13.2 is highly unlikely to play any role in the pathogenesis of sex reversal in this family as only two (III1 and III5) of the three affected siblings as well as an unaffected sister III4 had this particular CNV. Similarly, the 30kb duplication in 1p31.1 was excluded because it was present in all seven family members (both parents and five siblings). The 195kb deletion in 7q31.1 was found only in a normal 46,XX sibling III-4. Only the 349kb deletion segregated with the three affected siblings, the father from whom they inherited the deletion and was not detected in any unaffected family members.
Initially, SOX9 did not seem to be a prime candidate gene, as there were no bone deformities in the any of the affected individuals. However, the consistent deletions upstream of this gene in all three affected siblings led us to investigate the possibility that this could be the causative of sex reversal in this family. We confirmed the deletion by performing quantitative PCR on genomic DNA and demonstrating an approximately 50% reduction in the father and the three affected siblings. We hypothesized that mRNA expression of SOX9 in these subjects could be decreased compared with controls secondary to the deletion. RT-qPCR confirmed a statistically significant reduction in the expression of RNA in the three affected siblings.
Mutations in SOX9 gene usually result in campomelic dysplasia, which consists of short bowed lower extremities and other defects such as talipes equinovarus, congenital hip dislocation, mid-facial hypoplasia, dolichocephaly, and ambiguous genitalia. Familial cases have been described and ascribed to somatic mosaicism with mutations (7), whole gene deletion (24), and germ line mutation (25). It should be noted that all of these patients had campomelic dysplasia as well.
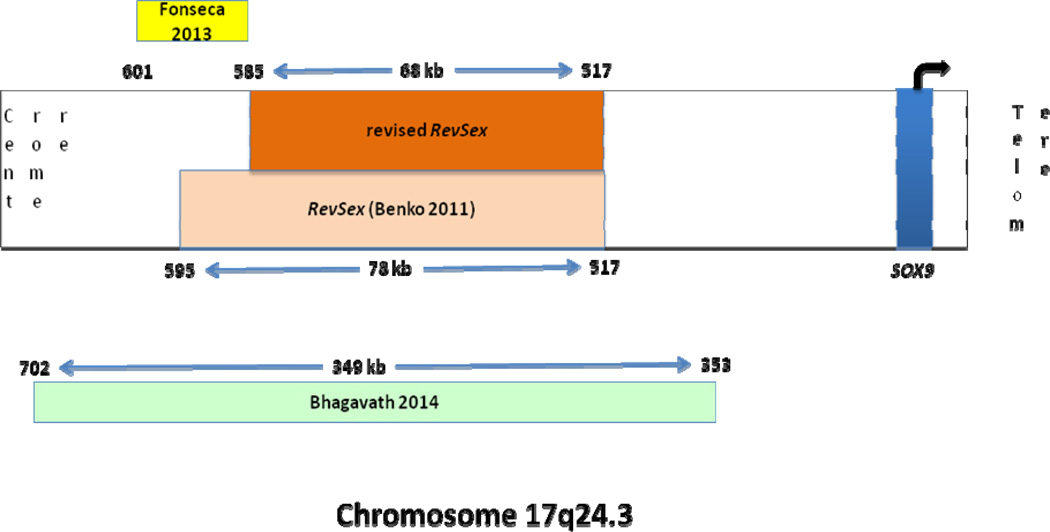
Pfeifer and coworkers had shown that deletions and translocations involving a region 140kb to 950 kb proximal to the SOX9 gene on chromosome 17 was the cause for campomelic dysplasia and XY sex reversal in three patients(26). After studying a 1.5 Mb deletion 380kb upstream of SOX9 in one patient, Pop and co-workers postulated that these rearrangements most likely involve one or more cis-regulatory elements (27). Of note, our 349 kb deletion upstream of SOX9 lies within this region.
Comparison of duplications in the upstream region of SOX9 in 46,XX sex reversed patients as well as the deletion in two 46,XY patients (cousins related by their mothers who were sisters) led Benko and co-workers to conclude that there is a specific, small region they called RevSex upstream of SOX9 (23). They identified this 78 kb region to be located 595 to 517 kb upstream of SOX9. They speculated based on comparison to other cases of larger duplications that involved much of the upstream region of SOX9, that a permissive ‘male’ chromatin environment is important for SOX9 activity with regards to development of the undifferentiated gonad into testis. This explained why the mothers with the deletion upstream of SOX9 had normal gonadal function whereas the affected children had streak gonads. They cite a similar description involving the Hoxd locus in support of their argument.
A more recent report of two patients with acampomelic campomelic dysplasia described balanced translocations involving chromosome 17 near the SOX9 gene. One of the patients had a normal male phenotype and the translocation break point was located within 601 to 585 kb upstream of SOX9 gene. This eliminates another 10 kb (595-585) from the RevSex region and further narrows it down to the region 585 to 517 kb upstream of SOX9 (Figure 6) (28). In support of Benko and co-workers (23), this specific RevSex region is contained in the deletion found in the three siblings and the father described in our study. In Benko’s cases, the deletions were maternally transmitted and the paternal allele of SOX9 gene was presumably still active. Since the mothers could not be affected, their ‘permissive chromatin environment’ theory seemed plausible. In contrast, in the family described in this study, the father had the same deletion and did not exhibit a sex-reversed phenotype whereas all three XY children exhibited the same sex reversal phenotype. The ‘permissive chromatin environment’ theory cannot explain why the father is not affected in this family. Alternative explanations were considered.
Figure 6.
Schematic representation of chromosome 17 with the position of SOX9 indicated relative to the centromere and telomere. The position of the original ‘RevSex’ region described by Benko et al is indicated within the chromosome. The numbers indicate the distance (in kb) upstream from SOX9. The position and size of the deletion described by Fonseca et al is shown outside the chromosome, above the schematic. The position and size of the deletion described in the current study is indicated below and outside the representation of the chromosome. Finally, the position and size of the ‘revised RevSex’ region based on the new deletions is shown within the representation of the chromosome above the box indicating the original ‘RevSex’ region.
Since SOX9 gene expression is decreased in the affected members of the family, we have to consider the possibility that a critical level of expression of SOX9 is needed for induction of the male sex-determining pathway. It is possible that although affected, the father did not have the critical reduction in SOX9 expression to be affected. The three affected siblings on the other hand may have decreased levels beneath this threshold and therefore have sex reversal. An alternative explanation is also possible and should be considered based on an elegant experiment performed in mice which showed that a combination of imprinting and maternal effect genes has control over expression of SOX9 genes (29). OdsSex mice show complete female-to-male sex reversal due to a tyrosinase transgene insertion 980kb upstream of Sox9. In contrast, if there is paternal transmission of the transgene combined with a maternal-effect locus, sex reversal can be suppressed in these mice.
We postulate that SOX9 expression in humans is also controlled in a complex manner, similar to that in mice as shown by Poirier and co-workers (29). This could explain why the father with the XY karyotype and the same 349 kb deletion at 17q24.3 located 353 kb upstream of SOX9 was not affected and did not develop 46,XY sex reversal. We hypothesize that he had a different allele from his mother that was able to overcome the effects of this deletion in one of his chromosomes. The three children on the other hand inherited the deletion from their father and might not have inherited the protective gene from their mother resulting in sex reversal. Imprinting effects, however, cannot be excluded.
Evidently, the SOX9 function essential for bone formation was not disrupted in the affected individuals in this study as there was no campomelic dysplasia. This supports the existence of proximal and distal campomelic dysplasia breakpoint clusters (which also happen to be proximal and distal to the SexRev region) involved in promotion of bone formation independent of the sex determination pathway(11). It is interesting that two of three affected sex reversed females had gonadoblastomas. These findings suggest that deletions upstream to SOX9 could predispose to gonadoblastoma and subsequent germ cell tumor risk. This will have to be confirmed in future studies.
5. CONCLUSION
Familial 46,XY sex reversal can be paternally inherited. To our knowledge this is the first report of a familial 46,XY sex reversal in three siblings caused by paternally inherited deletion in the region upstream of SOX9. Our report further confirms that a 67 kb region upstream of SOX9 gene is critical for the development of testis from the indeterminate gonad. Small deletions confined around this region can result in sex reversal without campomelic or acampomelic dysplasia and could predispose to gonadoblastoma.
HIGHLIGHTS.
Three siblings from a family of 5 sisters have 46, XY complete gonadal dysgenesis.
Deletion located upstream of the SOX9 gene in the father and the affected siblings.
Expression of SOX9 gene confirmed to be decreased in affected individuals.
Unique report of paternal inheritance of such a deletion with unaffected father.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Romao RL, Salle JL, Wherrett DK. Update on the management of disorders of sex development. Pediatr Clin North Am. 2012 Aug;59(4):853–869. doi: 10.1016/j.pcl.2012.05.020. [DOI] [PubMed] [Google Scholar]
- 2.Jager RJ, Anvret M, Hall K, Scherer G. A human XY female with a frame shift mutation in the candidate testis-determining gene SRY. Nature. 1990 Nov 29;348(6300):452–454. doi: 10.1038/348452a0. [DOI] [PubMed] [Google Scholar]
- 3.Assumpcao JG, Benedetti CE, Maciel-Guerra AT, Guerra G, Jr, Baptista MT, Scolfaro MR, et al. Novel mutations affecting SRY DNA-binding activity: The HMG box N65H associated with 46,XY pure gonadal dysgenesis and the familial non-HMG box R30I associated with variable phenotypes. J Mol Med (Berl) 2002 Dec;80(12):782–790. doi: 10.1007/s00109-002-0376-9. [DOI] [PubMed] [Google Scholar]
- 4.Ahmed SF, Bashamboo A, Lucas-Herald A, McElreavey K. Understanding the genetic aetiology in patients with XY DSD. Br Med Bull. 2013 Mar 25; doi: 10.1093/bmb/ldt008. [DOI] [PubMed] [Google Scholar]
- 5.Achermann JC, Ito M, Ito M, Hindmarsh PC, Jameson JL. A mutation in the gene encoding steroidogenic factor-1 causes XY sex reversal and adrenal failure in humans. Nat Genet. 1999 Jun;22(2):125–126. doi: 10.1038/9629. [DOI] [PubMed] [Google Scholar]
- 6.Sarafoglou K, Ostrer H. Clinical review 111: Familial sex reversal: A review. J Clin Endocrinol Metab. 2000 Feb;85(2):483–493. doi: 10.1210/jcem.85.2.6418. [DOI] [PubMed] [Google Scholar]
- 7.Wagner T, Wirth J, Meyer J, Zabel B, Held M, Zimmer J, et al. Autosomal sex reversal and campomelic dysplasia are caused by mutations in and around the SRY-related gene SOX9. Cell. 1994 Dec 16;79(6):1111–1120. doi: 10.1016/0092-8674(94)90041-8. [DOI] [PubMed] [Google Scholar]
- 8.Moog U, Jansen NJ, Scherer G, Schrander-Stumpel CT. Acampomelic campomelic syndrome. Am J Med Genet. 2001 Dec 1;104(3):239–245. [PubMed] [Google Scholar]
- 9.Bhagavath B, Xu N, Ozata M, Rosenfield RL, Bick DP, Sherins RJ, et al. KAL1 mutations are not a common cause of idiopathic hypogonadotrophic hypogonadism in humans. Mol Hum Reprod. 2007 Mar;13(3):165–170. doi: 10.1093/molehr/gal108. [DOI] [PubMed] [Google Scholar]
- 10.Lin L, Philibert P, Ferraz-de-Souza B, Kelberman D, Homfray T, Albanese A, et al. Heterozygous missense mutations in steroidogenic factor 1 (SF1/Ad4BP, NR5A1) are associated with 46,XY disorders of sex development with normal adrenal function. J Clin Endocrinol Metab. 2007 Mar;92(3):991–999. doi: 10.1210/jc.2006-1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miller DT, Shen Y, Wu BL. Oligonucleotide microarrays for clinical diagnosis of copy number variation and zygosity status. Curr Protoc Hum Genet. 2012 Jul; doi: 10.1002/0471142905.hg0812s74. Chapter 8 Unit8.12. [DOI] [PubMed] [Google Scholar]
- 12.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3(6):1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 13.Kutner M, Nachtsheim C, Neter J, Li W. Applied linear statistical models. 5th Edition ed. New York: McGraw-Hill; 2005. <br />. [Google Scholar]
- 14.Bryhn AC, Dimberg PH. An operational definition of a statistically meaningful trend. PLoS One. 2011 Apr 28;6(4):e19241. doi: 10.1371/journal.pone.0019241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hines RS, Tho SP, Zhang YY, Plouffe L, Jr, Hansen KA, Khan I, et al. Paternal somatic and germ-line mosaicism for a sex-determining region on Y (SRY) missense mutation leading to recurrent 46,XY sex reversal. Fertil Steril. 1997 Apr;67(4):675–679. doi: 10.1016/s0015-0282(97)81365-9. [DOI] [PubMed] [Google Scholar]
- 16.Saylam K, Simon P. WT1 gene mutation responsible for male sex reversal and renal failure: The frasier syndrome. Eur J Obstet Gynecol Reprod Biol. 2003 Sep 10;110(1):111–113. doi: 10.1016/s0301-2115(03)00088-5. [DOI] [PubMed] [Google Scholar]
- 17.Foster JW, Dominguez-Steglich MA, Guioli S, Kwok C, Weller PA, Stevanovic M, et al. Campomelic dysplasia and autosomal sex reversal caused by mutations in an SRY-related gene. Nature. 1994 Dec 8;372(6506):525–530. doi: 10.1038/372525a0. [DOI] [PubMed] [Google Scholar]
- 18.Lourenco D, Brauner R, Rybczynska M, Nihoul-Fekete C, McElreavey K, Bashamboo A. Loss-of-function mutation in GATA4 causes anomalies of human testicular development. Proc Natl Acad Sci U S A. 2011 Jan 25;108(4):1597–15602. doi: 10.1073/pnas.1010257108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liao X, Liang D, Li Y, Xi H, Quan Y, Wu L. Mutation analysis of the SRY, NR5A1, and DHH genes in six chinese 46,XY women. J Matern Fetal Neonatal Med. 2011 Jun;24(6):863–866. doi: 10.3109/14767058.2010.531321. [DOI] [PubMed] [Google Scholar]
- 20.Das DK, Sanghavi D, Gawde H, Idicula-Thomas S, Vasudevan L. Novel homozygous mutations in desert hedgehog gene in patients with 46,XY complete gonadal dysgenesis and prediction of its structural and functional implications by computational methods. Eur J Med Genet. 2011 Nov-Dec;54(6):e529–e534. doi: 10.1016/j.ejmg.2011.04.010. [DOI] [PubMed] [Google Scholar]
- 21.Biason-Lauber A, Konrad D, Meyer M, DeBeaufort C, Schoenle EJ. Ovaries and female phenotype in a girl with 46,XY karyotype and mutations in the CBX2 gene. Am J Hum Genet. 2009 May;84(5):658–663. doi: 10.1016/j.ajhg.2009.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ion A, Telvi L, Chaussain JL, Galacteros F, Valayer J, Fellous M, et al. A novel mutation in the putative DNA helicase XH2 is responsible for male-to-female sex reversal associated with an atypical form of the ATR-X syndrome. Am J Hum Genet. 1996 Jun;58(6):1185–1191. [PMC free article] [PubMed] [Google Scholar]
- 23.Benko S, Gordon CT, Mallet D, Sreenivasan R, Thauvin-Robinet C, Brendehaug A, et al. Disruption of a long distance regulatory region upstream of SOX9 in isolated disorders of sex development. J Med Genet. 2011 Dec;48(12):825–830. doi: 10.1136/jmedgenet-2011-100255. [DOI] [PubMed] [Google Scholar]
- 24.Smyk M, Obersztyn E, Nowakowska B, Bocian E, Cheung SW, Mazurczak T, et al. Recurrent SOX9 deletion campomelic dysplasia due to somatic mosaicism in the father. Am J Med Genet A. 2007 Apr 15;143A(8):866–870. doi: 10.1002/ajmg.a.31631. [DOI] [PubMed] [Google Scholar]
- 25.Cameron FJ, Hageman RM, Cooke-Yarborough C, Kwok C, Goodwin LL, Sillence DO, et al. A novel germ line mutation in SOX9 causes familial campomelic dysplasia and sex reversal. Hum Mol Genet. 1996 Oct;5(10):1625–1630. doi: 10.1093/hmg/5.10.1625. [DOI] [PubMed] [Google Scholar]
- 26.Pfeifer D, Kist R, Dewar K, Devon K, Lander ES, Birren B, et al. Campomelic dysplasia translocation breakpoints are scattered over 1 mb proximal to SOX9: Evidence for an extended control region. Am J Hum Genet. 1999 Jul;65(1):111–124. doi: 10.1086/302455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pop R, Conz C, Lindenberg KS, Blesson S, Schmalenberger B, Briault S, et al. Screening of the 1 mb SOX9 5' control region by array CGH identifies a large deletion in a case of campomelic dysplasia with XY sex reversal. J Med Genet. 2004 Apr;41(4):e47. doi: 10.1136/jmg.2003.013185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fonseca AC, Bonaldi A, Bertola DR, Kim CA, Otto PA, Vianna-Morgante AM. The clinical impact of chromosomal rearrangements with breakpoints upstream of the SOX9 gene: Two novel de novo balanced translocations associated with acampomelic campomelic dysplasia. BMC Med Genet. 2013 May 7;14(50) doi: 10.1186/1471-2350-14-50. 2350-14-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poirier C, Qin Y, Adams CP, Anaya Y, Singer JB, Hill AE, et al. A complex interaction of imprinted and maternal-effect genes modifies sex determination in odd sex (ods) mice. Genetics. 2004 Nov;168(3):1557–1562. doi: 10.1534/genetics.104.032177. 2004 Nov. [DOI] [PMC free article] [PubMed] [Google Scholar]