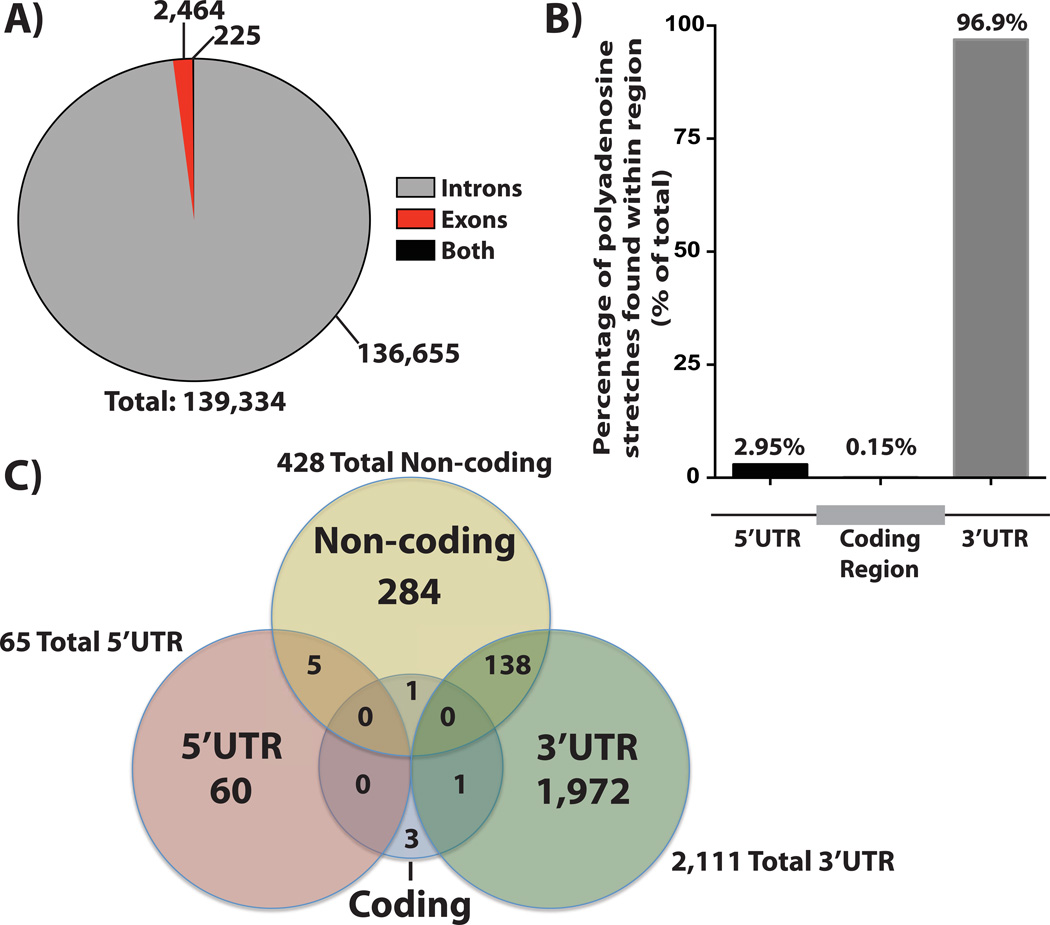
Figure 5. Prevalence and Location of Templated, Internal Polyadenosine Stretches Within the Human Transcriptome.
A) Analysis of the human transcriptome for the frequency and enrichment of internal polyadenosine stretches containing at least 12 consecutive adenosines reveals that the vast majority (136,655 out of 139,334) of these sequences are located in the introns of mRNAs whereas a much smaller fraction are located in exonic regions (2,464 out of 139,334), which includes 5’ and 3’UTR regions. A much smaller number of these internal adenosine stretches are found in sequences that can be either introns or exons as a result of alternative splice variants. B) Internal polyadenosine stretches found in exonic sequences are almost exclusively located in untranslated regions. Further analysis of the ≥12nt polyadenosine sequences that occur in mature mRNA transcripts reveals that almost all (99.9%) of these instances are located in the UTRs, with an extremely large percentage present in the 3’UTRs (96.9%) of mRNA transcripts. C) Non-coding RNAs also include templated stretches of polyadenosine. A number (428) of noncoding RNAs (yellow circle) contain templated polyadenosine sequences, suggesting that Pabs could modulate non-coding RNAs through this templated binding site. Consistent with a model of 3’ UTR regulation, a large number of the internal polyadenosine stretches identified in ncRNAs (138) are transcribed from regions that correspond to the 3’UTRs of other/host genes (yellow and green overlap).